Cameron Musco
No-regret Algorithms for Fair Resource Allocation
Mar 11, 2023



Abstract:We consider a fair resource allocation problem in the no-regret setting against an unrestricted adversary. The objective is to allocate resources equitably among several agents in an online fashion so that the difference of the aggregate $\alpha$-fair utilities of the agents between an optimal static clairvoyant allocation and that of the online policy grows sub-linearly with time. The problem is challenging due to the non-additive nature of the $\alpha$-fairness function. Previously, it was shown that no online policy can exist for this problem with a sublinear standard regret. In this paper, we propose an efficient online resource allocation policy, called Online Proportional Fair (OPF), that achieves $c_\alpha$-approximate sublinear regret with the approximation factor $c_\alpha=(1-\alpha)^{-(1-\alpha)}\leq 1.445,$ for $0\leq \alpha < 1$. The upper bound to the $c_\alpha$-regret for this problem exhibits a surprising phase transition phenomenon. The regret bound changes from a power-law to a constant at the critical exponent $\alpha=\frac{1}{2}.$ As a corollary, our result also resolves an open problem raised by Even-Dar et al. [2009] on designing an efficient no-regret policy for the online job scheduling problem in certain parameter regimes. The proof of our results introduces new algorithmic and analytical techniques, including greedy estimation of the future gradients for non-additive global reward functions and bootstrapping adaptive regret bounds, which may be of independent interest.
Sample Constrained Treatment Effect Estimation
Oct 12, 2022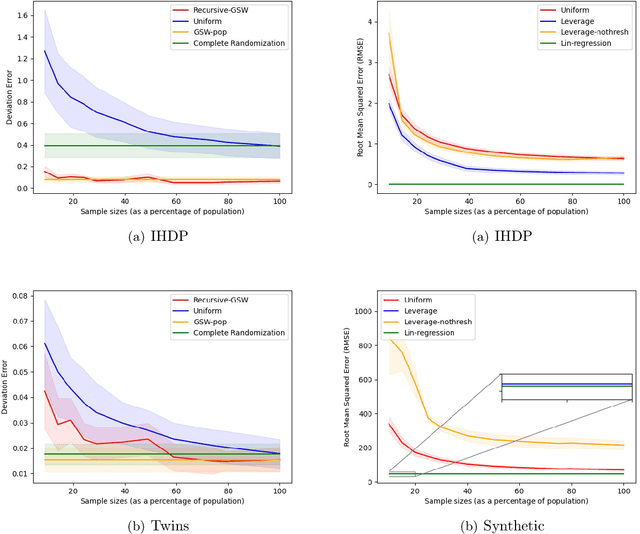
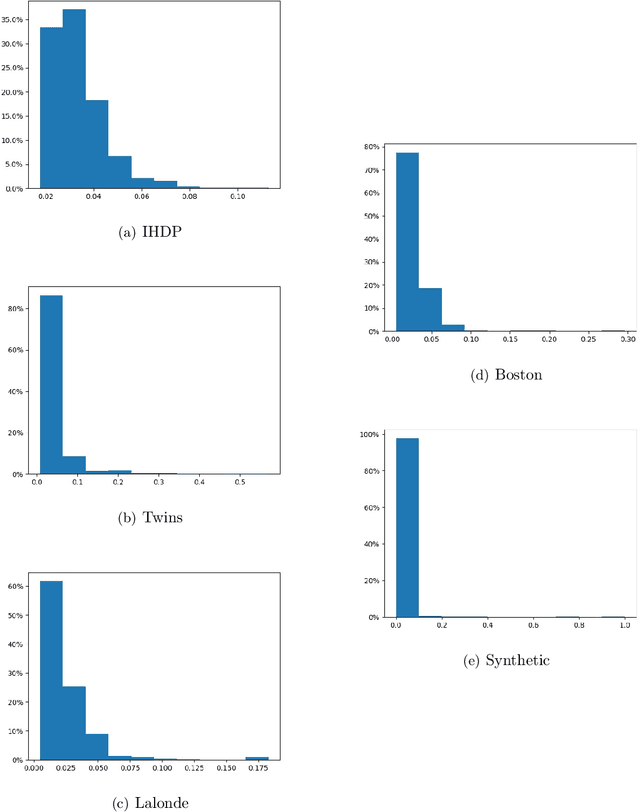
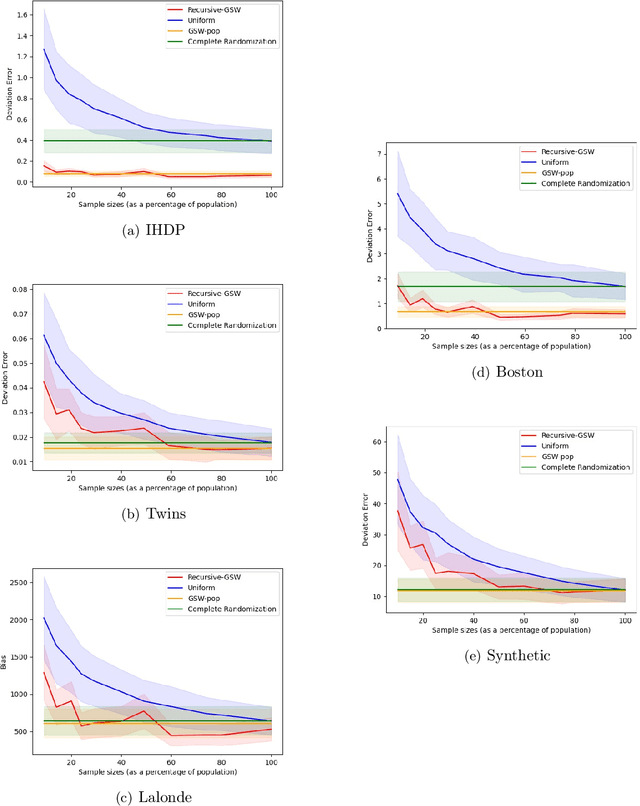
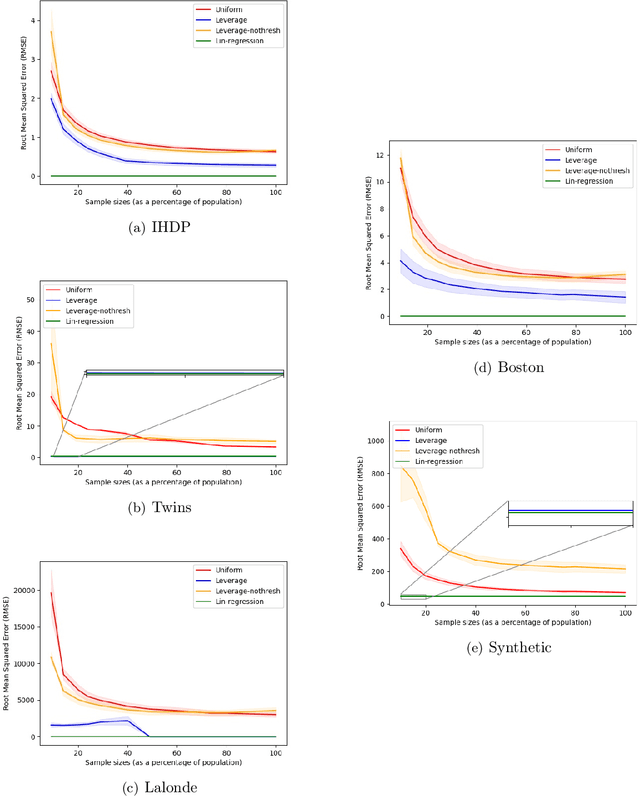
Abstract:Treatment effect estimation is a fundamental problem in causal inference. We focus on designing efficient randomized controlled trials, to accurately estimate the effect of some treatment on a population of $n$ individuals. In particular, we study sample-constrained treatment effect estimation, where we must select a subset of $s \ll n$ individuals from the population to experiment on. This subset must be further partitioned into treatment and control groups. Algorithms for partitioning the entire population into treatment and control groups, or for choosing a single representative subset, have been well-studied. The key challenge in our setting is jointly choosing a representative subset and a partition for that set. We focus on both individual and average treatment effect estimation, under a linear effects model. We give provably efficient experimental designs and corresponding estimators, by identifying connections to discrepancy minimization and leverage-score-based sampling used in randomized numerical linear algebra. Our theoretical results obtain a smooth transition to known guarantees when $s$ equals the population size. We also empirically demonstrate the performance of our algorithms.
Direct Embedding of Temporal Network Edges via Time-Decayed Line Graphs
Sep 30, 2022
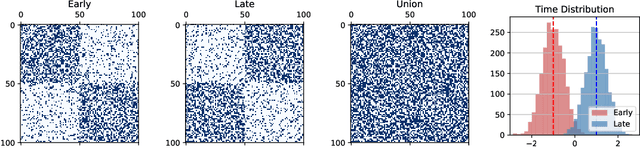
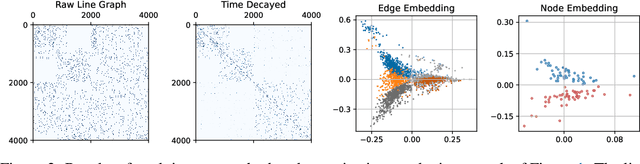
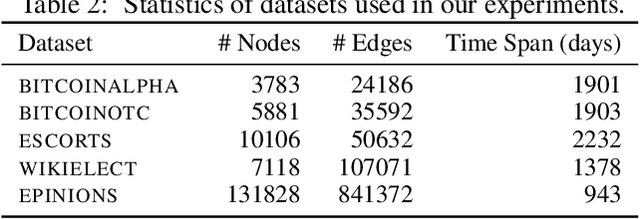
Abstract:Temporal networks model a variety of important phenomena involving timed interactions between entities. Existing methods for machine learning on temporal networks generally exhibit at least one of two limitations. First, time is assumed to be discretized, so if the time data is continuous, the user must determine the discretization and discard precise time information. Second, edge representations can only be calculated indirectly from the nodes, which may be suboptimal for tasks like edge classification. We present a simple method that avoids both shortcomings: construct the line graph of the network, which includes a node for each interaction, and weigh the edges of this graph based on the difference in time between interactions. From this derived graph, edge representations for the original network can be computed with efficient classical methods. The simplicity of this approach facilitates explicit theoretical analysis: we can constructively show the effectiveness of our method's representations for a natural synthetic model of temporal networks. Empirical results on real-world networks demonstrate our method's efficacy and efficiency on both edge classification and temporal link prediction.
Simplified Graph Convolution with Heterophily
Feb 08, 2022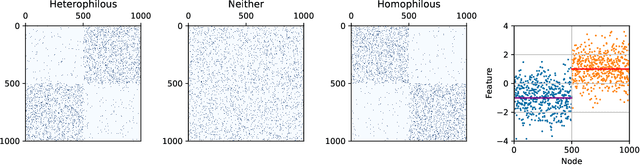
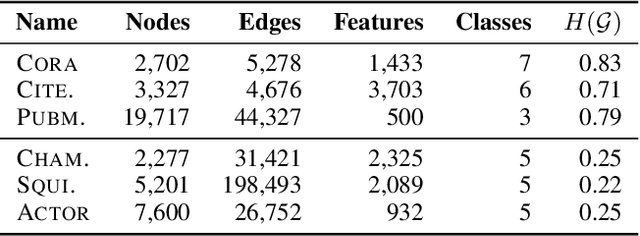
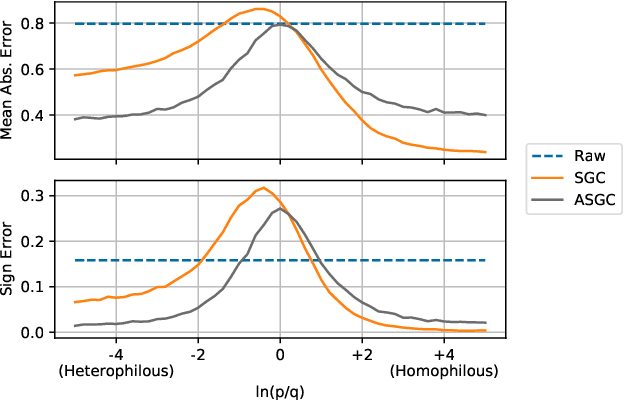
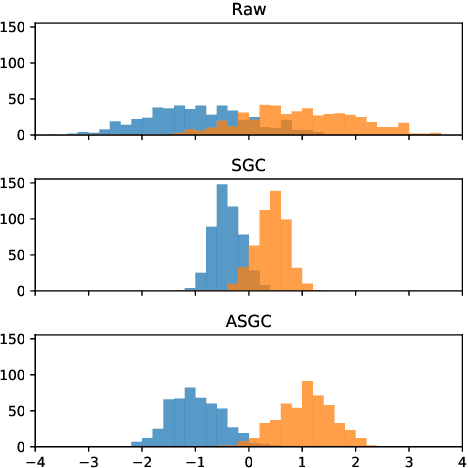
Abstract:Graph convolutional networks (GCNs) (Kipf & Welling, 2017) attempt to extend the success of deep learning in modeling image and text data to graphs. However, like other deep models, GCNs comprise repeated nonlinear transformations of inputs and are therefore time and memory intensive to train. Recent work has shown that a much simpler and faster model, Simple Graph Convolution (SGC) (Wu et al., 2019), is competitive with GCNs in common graph machine learning benchmarks. The use of graph data in SGC implicitly assumes the common but not universal graph characteristic of homophily, wherein nodes link to nodes which are similar. Here we show that SGC is indeed ineffective for heterophilous (i.e., non-homophilous) graphs via experiments on synthetic and real-world datasets. We propose Adaptive Simple Graph Convolution (ASGC), which we show can adapt to both homophilous and heterophilous graph structure. Like SGC, ASGC is not a deep model, and hence is fast, scalable, and interpretable. We find that our non-deep method often outperforms state-of-the-art deep models at node classification on a benchmark of real-world datasets. The SGC paper questioned whether the complexity of graph neural networks is warranted for common graph problems involving homophilous networks; our results suggest that this question is still open even for more complicated problems involving heterophilous networks.
Sublinear Time Approximation of Text Similarity Matrices
Dec 17, 2021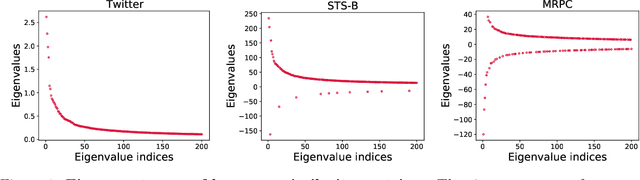
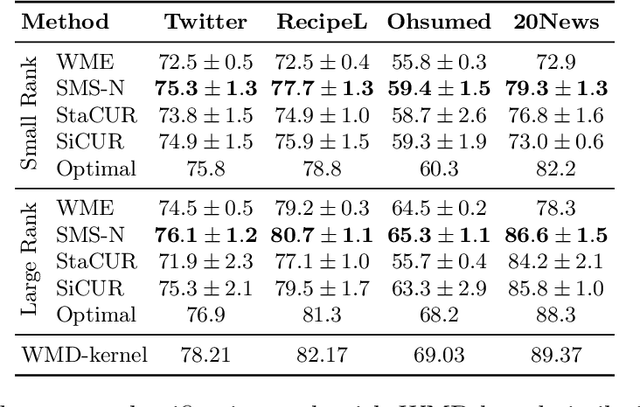
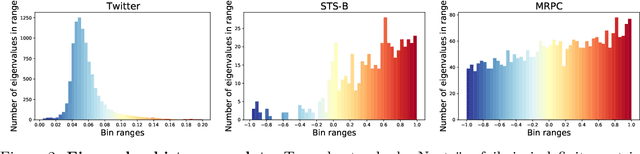
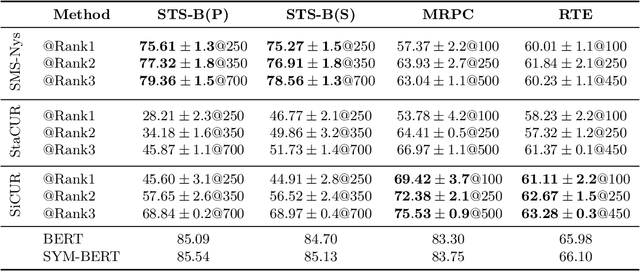
Abstract:We study algorithms for approximating pairwise similarity matrices that arise in natural language processing. Generally, computing a similarity matrix for $n$ data points requires $\Omega(n^2)$ similarity computations. This quadratic scaling is a significant bottleneck, especially when similarities are computed via expensive functions, e.g., via transformer models. Approximation methods reduce this quadratic complexity, often by using a small subset of exactly computed similarities to approximate the remainder of the complete pairwise similarity matrix. Significant work focuses on the efficient approximation of positive semidefinite (PSD) similarity matrices, which arise e.g., in kernel methods. However, much less is understood about indefinite (non-PSD) similarity matrices, which often arise in NLP. Motivated by the observation that many of these matrices are still somewhat close to PSD, we introduce a generalization of the popular Nystr\"{o}m method to the indefinite setting. Our algorithm can be applied to any similarity matrix and runs in sublinear time in the size of the matrix, producing a rank-$s$ approximation with just $O(ns)$ similarity computations. We show that our method, along with a simple variant of CUR decomposition, performs very well in approximating a variety of similarity matrices arising in NLP tasks. We demonstrate high accuracy of the approximated similarity matrices in the downstream tasks of document classification, sentence similarity, and cross-document coreference.
Active Sampling for Linear Regression Beyond the $\ell_2$ Norm
Nov 09, 2021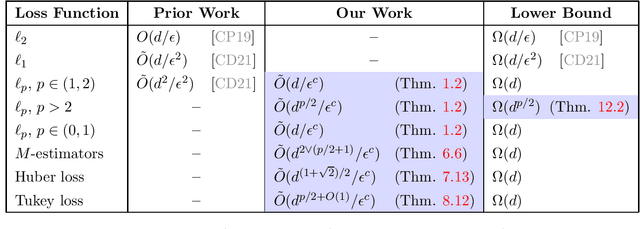
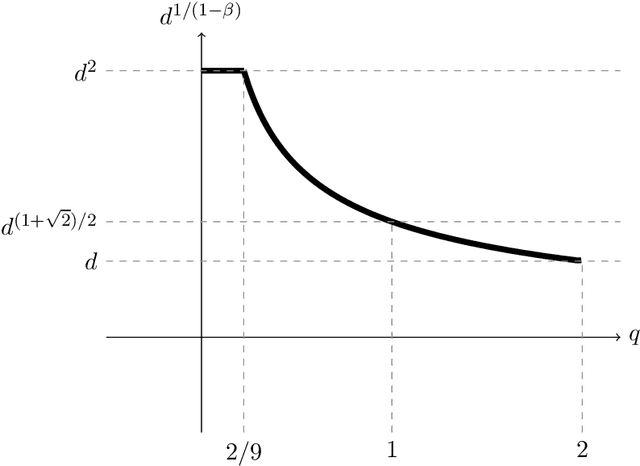
Abstract:We study active sampling algorithms for linear regression, which aim to query only a small number of entries of a target vector $b\in\mathbb{R}^n$ and output a near minimizer to $\min_{x\in\mathbb{R}^d}\|Ax-b\|$, where $A\in\mathbb{R}^{n \times d}$ is a design matrix and $\|\cdot\|$ is some loss function. For $\ell_p$ norm regression for any $0<p<\infty$, we give an algorithm based on Lewis weight sampling that outputs a $(1+\epsilon)$ approximate solution using just $\tilde{O}(d^{\max(1,{p/2})}/\mathrm{poly}(\epsilon))$ queries to $b$. We show that this dependence on $d$ is optimal, up to logarithmic factors. Our result resolves a recent open question of Chen and Derezi\'{n}ski, who gave near optimal bounds for the $\ell_1$ norm, and suboptimal bounds for $\ell_p$ regression with $p\in(1,2)$. We also provide the first total sensitivity upper bound of $O(d^{\max\{1,p/2\}}\log^2 n)$ for loss functions with at most degree $p$ polynomial growth. This improves a recent result of Tukan, Maalouf, and Feldman. By combining this with our techniques for the $\ell_p$ regression result, we obtain an active regression algorithm making $\tilde O(d^{1+\max\{1,p/2\}}/\mathrm{poly}(\epsilon))$ queries, answering another open question of Chen and Derezi\'{n}ski. For the important special case of the Huber loss, we further improve our bound to an active sample complexity of $\tilde O(d^{(1+\sqrt2)/2}/\epsilon^c)$ and a non-active sample complexity of $\tilde O(d^{4-2\sqrt 2}/\epsilon^c)$, improving a previous $d^4$ bound for Huber regression due to Clarkson and Woodruff. Our sensitivity bounds have further implications, improving a variety of previous results using sensitivity sampling, including Orlicz norm subspace embeddings and robust subspace approximation. Finally, our active sampling results give the first sublinear time algorithms for Kronecker product regression under every $\ell_p$ norm.
An Interpretable Graph Generative Model with Heterophily
Nov 04, 2021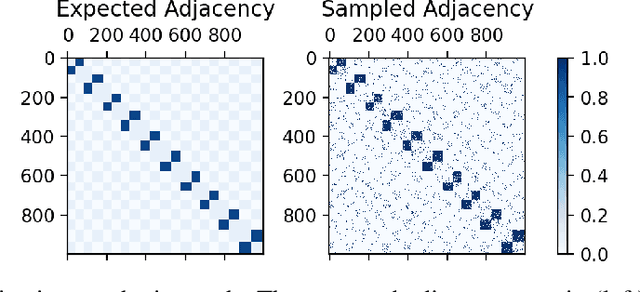
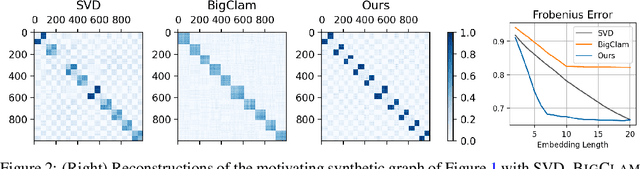
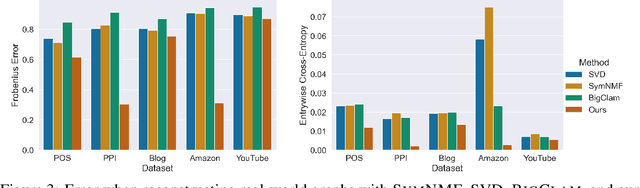
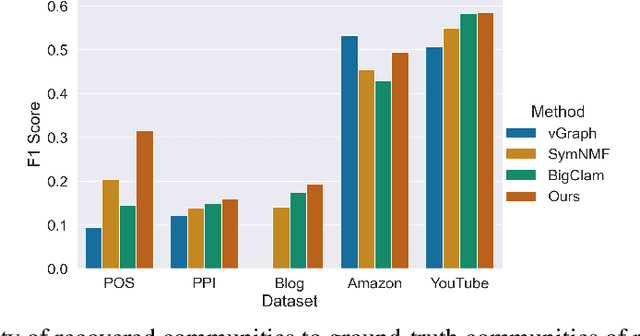
Abstract:Many models for graphs fall under the framework of edge-independent dot product models. These models output the probabilities of edges existing between all pairs of nodes, and the probability of a link between two nodes increases with the dot product of vectors associated with the nodes. Recent work has shown that these models are unable to capture key structures in real-world graphs, particularly heterophilous structures, wherein links occur between dissimilar nodes. We propose the first edge-independent graph generative model that is a) expressive enough to capture heterophily, b) produces nonnegative embeddings, which allow link predictions to be interpreted in terms of communities, and c) optimizes effectively on real-world graphs with gradient descent on a cross-entropy loss. Our theoretical results demonstrate the expressiveness of our model in its ability to exactly reconstruct a graph using a number of clusters that is linear in the maximum degree, along with its ability to capture both heterophily and homophily in the data. Further, our experiments demonstrate the effectiveness of our model for a variety of important application tasks such as multi-label clustering and link prediction.
On the Power of Edge Independent Graph Models
Oct 29, 2021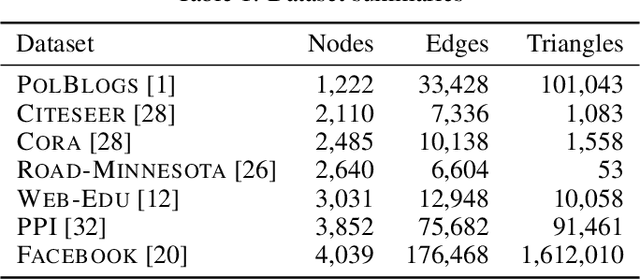
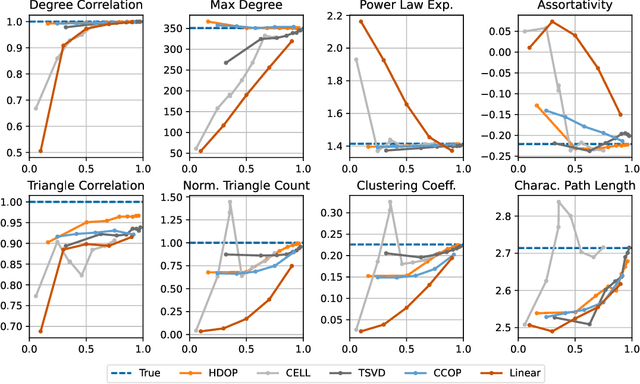
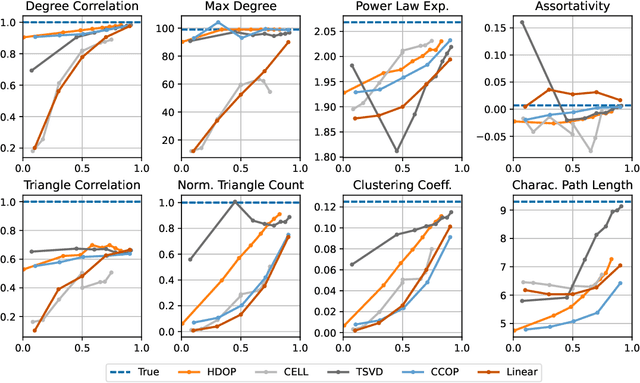
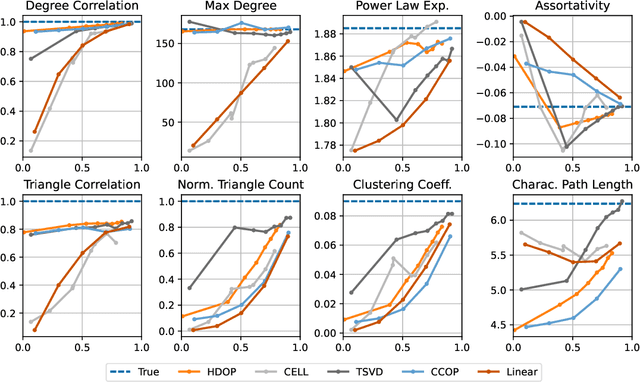
Abstract:Why do many modern neural-network-based graph generative models fail to reproduce typical real-world network characteristics, such as high triangle density? In this work we study the limitations of edge independent random graph models, in which each edge is added to the graph independently with some probability. Such models include both the classic Erd\"{o}s-R\'{e}nyi and stochastic block models, as well as modern generative models such as NetGAN, variational graph autoencoders, and CELL. We prove that subject to a bounded overlap condition, which ensures that the model does not simply memorize a single graph, edge independent models are inherently limited in their ability to generate graphs with high triangle and other subgraph densities. Notably, such high densities are known to appear in real-world social networks and other graphs. We complement our negative results with a simple generative model that balances overlap and accuracy, performing comparably to more complex models in reconstructing many graph statistics.
Coresets for Classification -- Simplified and Strengthened
Jun 18, 2021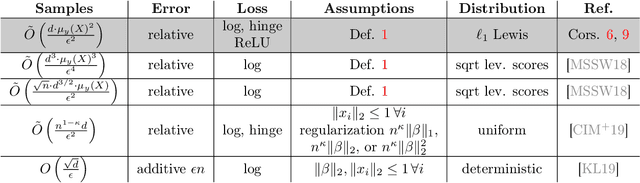
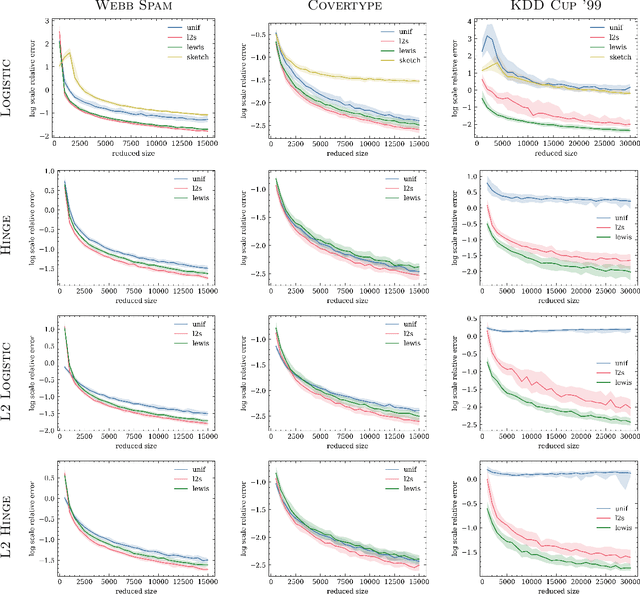
Abstract:We give relative error coresets for training linear classifiers with a broad class of loss functions, including the logistic loss and hinge loss. Our construction achieves $(1\pm \epsilon)$ relative error with $\tilde O(d \cdot \mu_y(X)^2/\epsilon^2)$ points, where $\mu_y(X)$ is a natural complexity measure of the data matrix $X \in \mathbb{R}^{n \times d}$ and label vector $y \in \{-1,1\}^n$, introduced in by Munteanu et al. 2018. Our result is based on subsampling data points with probabilities proportional to their $\ell_1$ $Lewis$ $weights$. It significantly improves on existing theoretical bounds and performs well in practice, outperforming uniform subsampling along with other importance sampling methods. Our sampling distribution does not depend on the labels, so can be used for active learning. It also does not depend on the specific loss function, so a single coreset can be used in multiple training scenarios.
DeepWalking Backwards: From Embeddings Back to Graphs
Feb 17, 2021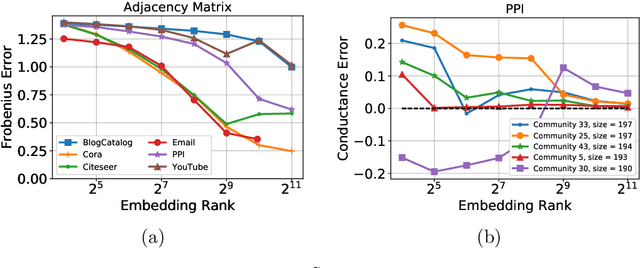
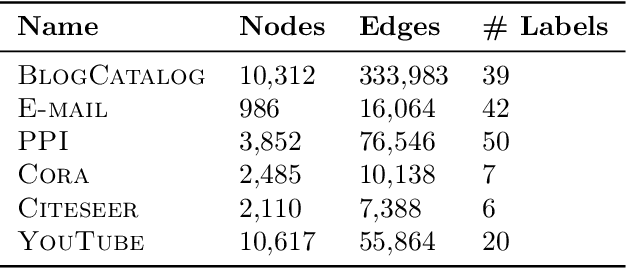
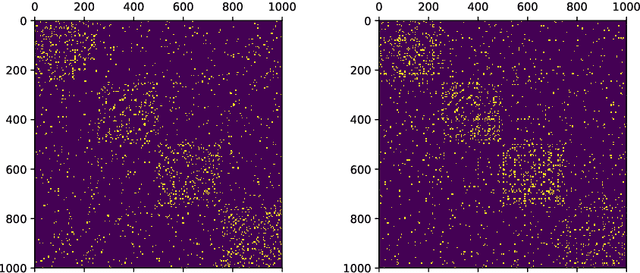
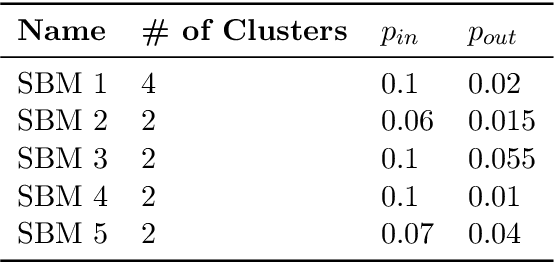
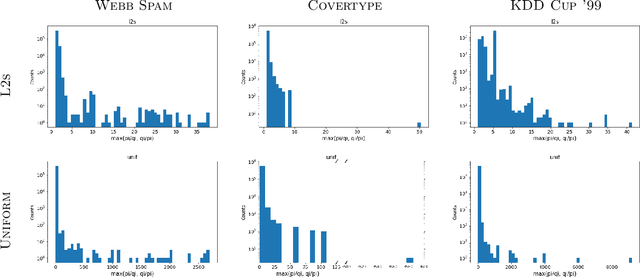
Abstract:Low-dimensional node embeddings play a key role in analyzing graph datasets. However, little work studies exactly what information is encoded by popular embedding methods, and how this information correlates with performance in downstream machine learning tasks. We tackle this question by studying whether embeddings can be inverted to (approximately) recover the graph used to generate them. Focusing on a variant of the popular DeepWalk method (Perozzi et al., 2014; Qiu et al., 2018), we present algorithms for accurate embedding inversion - i.e., from the low-dimensional embedding of a graph G, we can find a graph H with a very similar embedding. We perform numerous experiments on real-world networks, observing that significant information about G, such as specific edges and bulk properties like triangle density, is often lost in H. However, community structure is often preserved or even enhanced. Our findings are a step towards a more rigorous understanding of exactly what information embeddings encode about the input graph, and why this information is useful for learning tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge