Zhiqiang Hu
Multi-frame Collaboration for Effective Endoscopic Video Polyp Detection via Spatial-Temporal Feature Transformation
Jul 08, 2021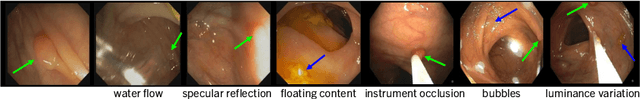
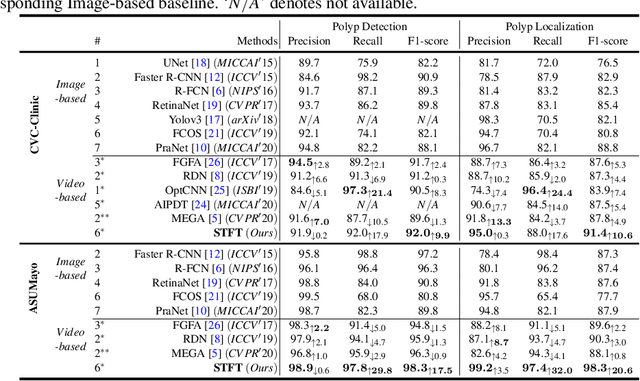
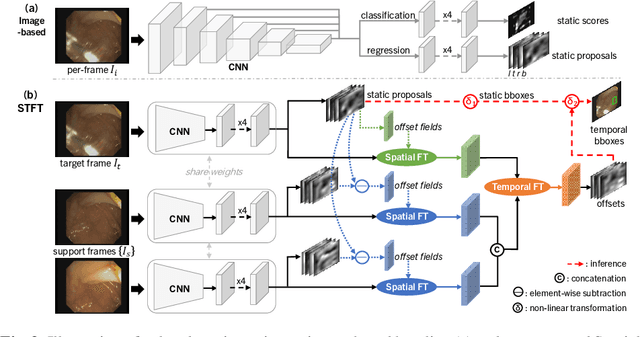

Abstract:Precise localization of polyp is crucial for early cancer screening in gastrointestinal endoscopy. Videos given by endoscopy bring both richer contextual information as well as more challenges than still images. The camera-moving situation, instead of the common camera-fixed-object-moving one, leads to significant background variation between frames. Severe internal artifacts (e.g. water flow in the human body, specular reflection by tissues) can make the quality of adjacent frames vary considerately. These factors hinder a video-based model to effectively aggregate features from neighborhood frames and give better predictions. In this paper, we present Spatial-Temporal Feature Transformation (STFT), a multi-frame collaborative framework to address these issues. Spatially, STFT mitigates inter-frame variations in the camera-moving situation with feature alignment by proposal-guided deformable convolutions. Temporally, STFT proposes a channel-aware attention module to simultaneously estimate the quality and correlation of adjacent frames for adaptive feature aggregation. Empirical studies and superior results demonstrate the effectiveness and stability of our method. For example, STFT improves the still image baseline FCOS by 10.6% and 20.6% on the comprehensive F1-score of the polyp localization task in CVC-Clinic and ASUMayo datasets, respectively, and outperforms the state-of-the-art video-based method by 3.6% and 8.0%, respectively. Code is available at \url{https://github.com/lingyunwu14/STFT}.
Mixed Supervision Learning for Whole Slide Image Classification
Jul 05, 2021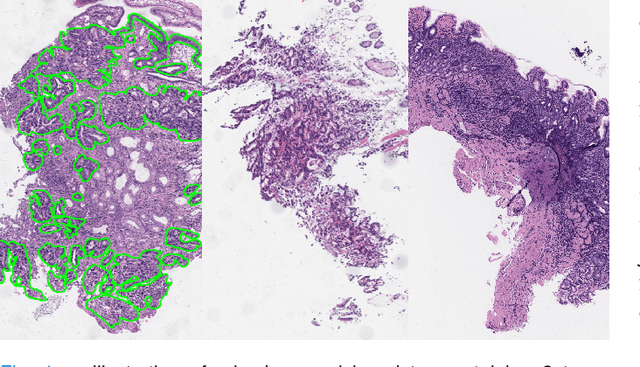
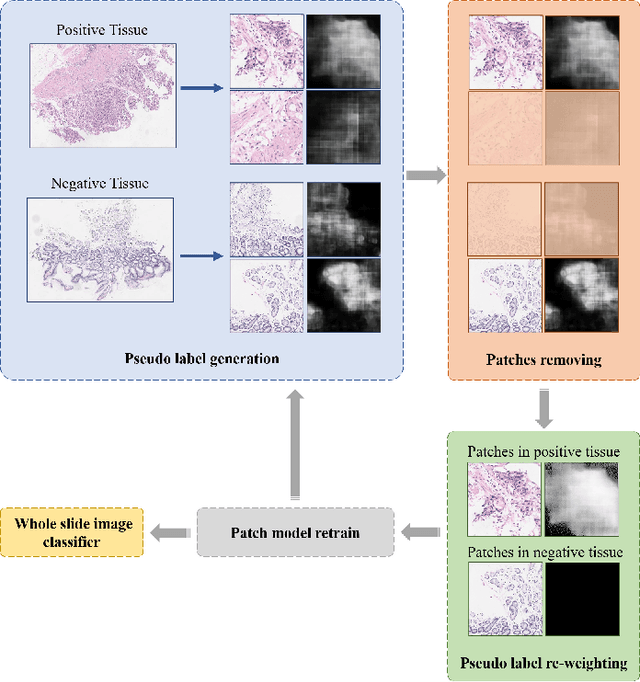
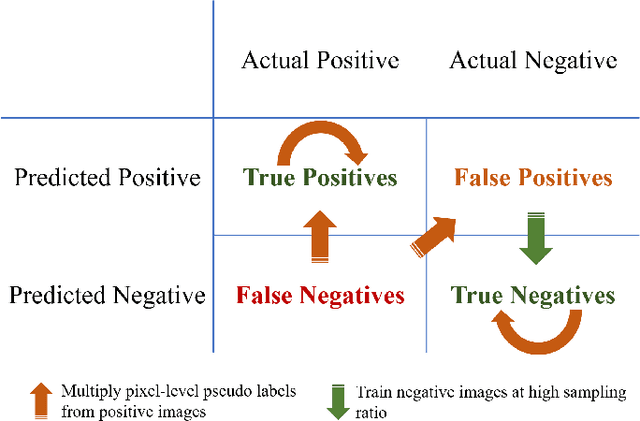
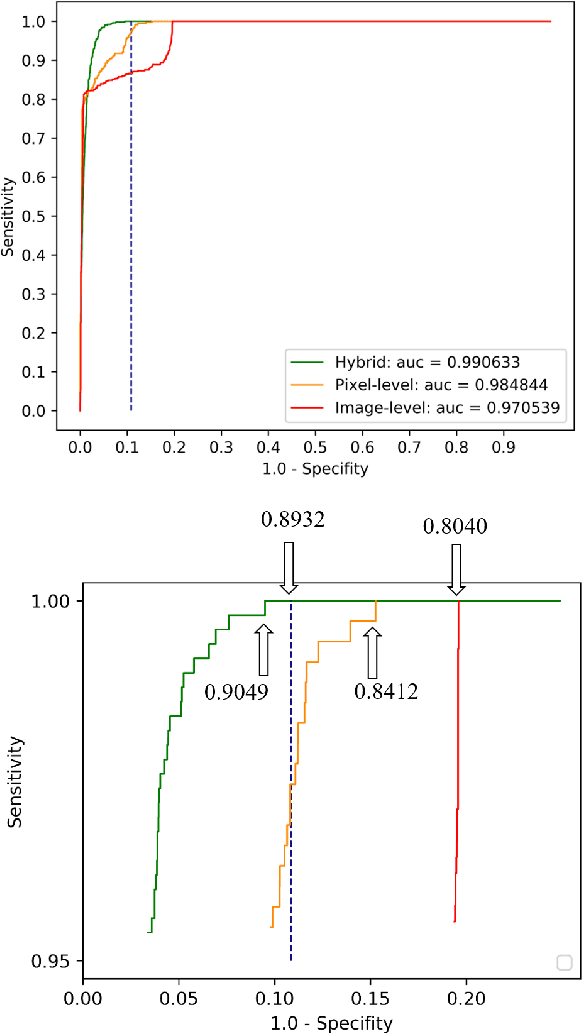
Abstract:Weak supervision learning on classification labels has demonstrated high performance in various tasks. When a few pixel-level fine annotations are also affordable, it is natural to leverage both of the pixel-level (e.g., segmentation) and image level (e.g., classification) annotation to further improve the performance. In computational pathology, however, such weak or mixed supervision learning is still a challenging task, since the high resolution of whole slide images makes it unattainable to perform end-to-end training of classification models. An alternative approach is to analyze such data by patch-base model training, i.e., using self-supervised learning to generate pixel-level pseudo labels for patches. However, such methods usually have model drifting issues, i.e., hard to converge, because the noise accumulates during the self-training process. To handle those problems, we propose a mixed supervision learning framework for super high-resolution images to effectively utilize their various labels (e.g., sufficient image-level coarse annotations and a few pixel-level fine labels). During the patch training stage, this framework can make use of coarse image-level labels to refine self-supervised learning and generate high-quality pixel-level pseudo labels. A comprehensive strategy is proposed to suppress pixel-level false positives and false negatives. Three real-world datasets with very large number of images (i.e., more than 10,000 whole slide images) and various types of labels are used to evaluate the effectiveness of mixed supervision learning. We reduced the false positive rate by around one third compared to state of the art while retaining 100% sensitivity, in the task of image-level classification.
Learning Unknown from Correlations: Graph Neural Network for Inter-novel-protein Interaction Prediction
Jun 01, 2021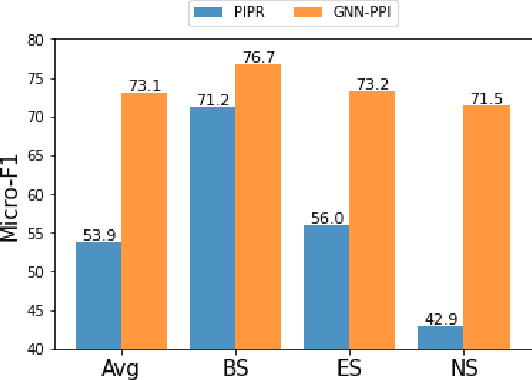
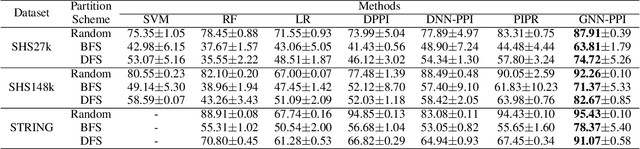
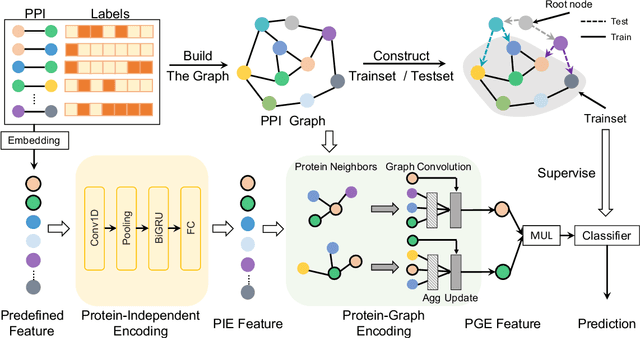
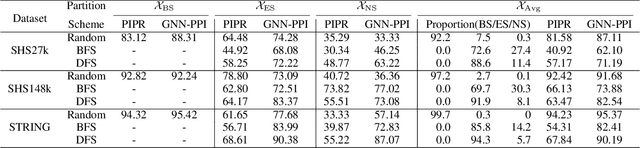
Abstract:The study of multi-type Protein-Protein Interaction (PPI) is fundamental for understanding biological processes from a systematic perspective and revealing disease mechanisms. Existing methods suffer from significant performance degradation when tested in unseen dataset. In this paper, we investigate the problem and find that it is mainly attributed to the poor performance for inter-novel-protein interaction prediction. However, current evaluations overlook the inter-novel-protein interactions, and thus fail to give an instructive assessment. As a result, we propose to address the problem from both the evaluation and the methodology. Firstly, we design a new evaluation framework that fully respects the inter-novel-protein interactions and gives consistent assessment across datasets. Secondly, we argue that correlations between proteins must provide useful information for analysis of novel proteins, and based on this, we propose a graph neural network based method (GNN-PPI) for better inter-novel-protein interaction prediction. Experimental results on real-world datasets of different scales demonstrate that GNN-PPI significantly outperforms state-of-the-art PPI prediction methods, especially for the inter-novel-protein interaction prediction.
DeepStyle: User Style Embedding for Authorship Attribution of Short Texts
Mar 14, 2021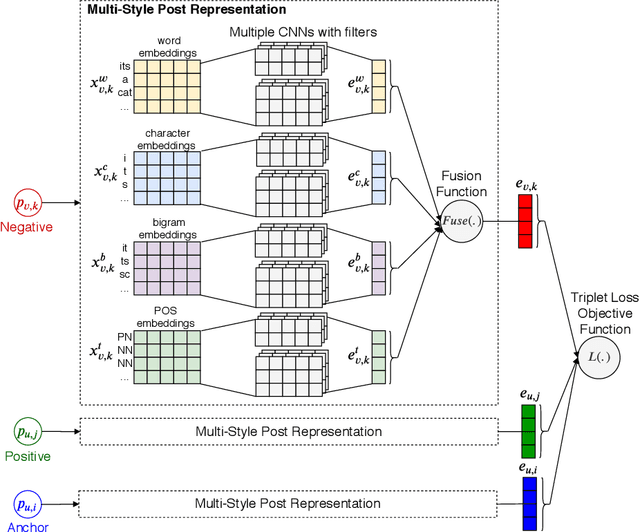
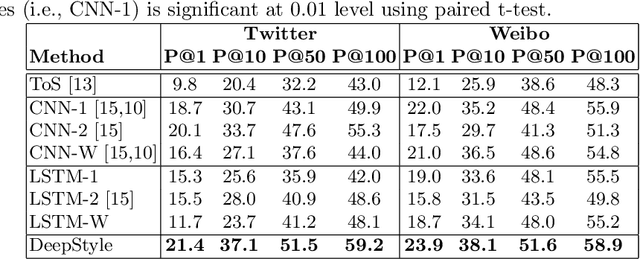
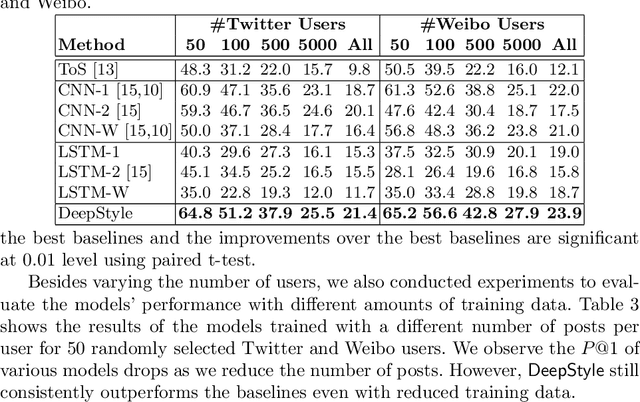
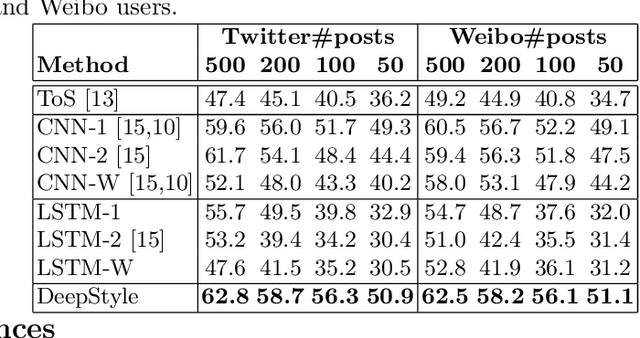
Abstract:Authorship attribution (AA), which is the task of finding the owner of a given text, is an important and widely studied research topic with many applications. Recent works have shown that deep learning methods could achieve significant accuracy improvement for the AA task. Nevertheless, most of these proposed methods represent user posts using a single type of feature (e.g., word bi-grams) and adopt a text classification approach to address the task. Furthermore, these methods offer very limited explainability of the AA results. In this paper, we address these limitations by proposing DeepStyle, a novel embedding-based framework that learns the representations of users' salient writing styles. We conduct extensive experiments on two real-world datasets from Twitter and Weibo. Our experiment results show that DeepStyle outperforms the state-of-the-art baselines on the AA task.
Text Style Transfer: A Review and Experiment Evaluation
Oct 24, 2020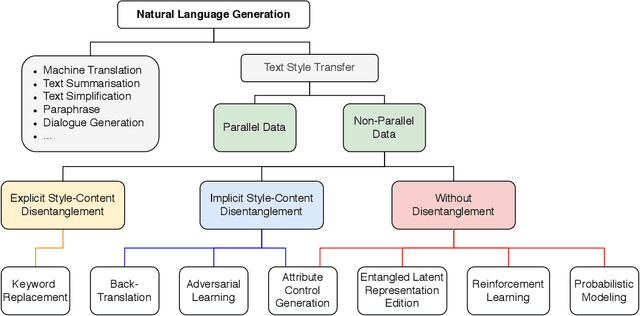
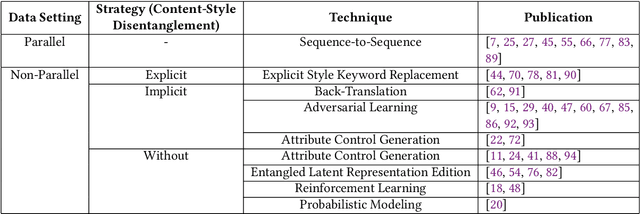
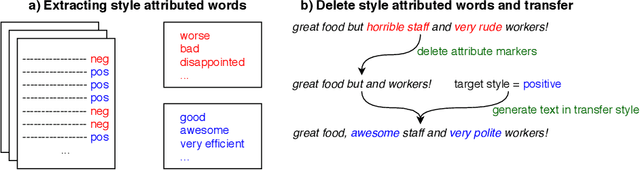
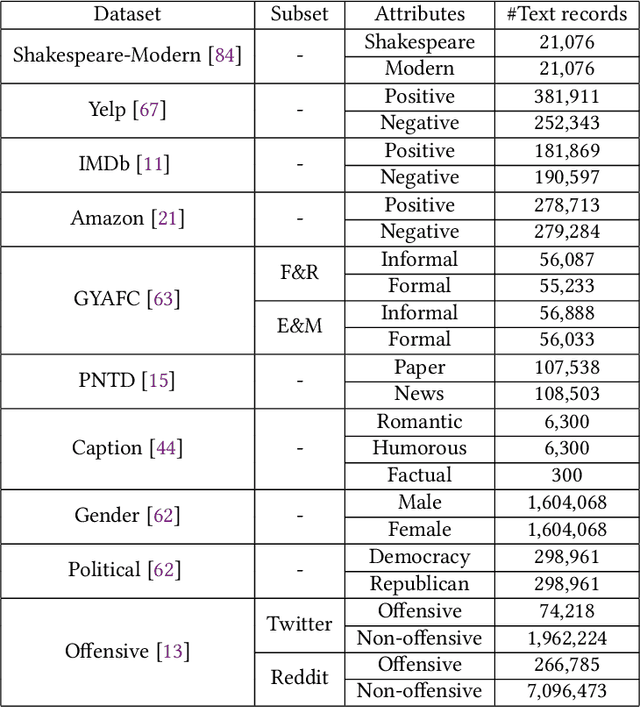
Abstract:The stylistic properties of text have intrigued computational linguistics researchers in recent years. Specifically, researchers have investigated the Text Style Transfer (TST) task, which aims to change the stylistic properties of the text while retaining its style independent content. Over the last few years, many novel TST algorithms have been developed, while the industry has leveraged these algorithms to enable exciting TST applications. The field of TST research has burgeoned because of this symbiosis. This article aims to provide a comprehensive review of recent research efforts on text style transfer. More concretely, we create a taxonomy to organize the TST models and provide a comprehensive summary of the state of the art. We review the existing evaluation methodologies for TST tasks and conduct a large-scale reproducibility study where we experimentally benchmark 19 state-of-the-art TST algorithms on two publicly available datasets. Finally, we expand on current trends and provide new perspectives on the new and exciting developments in the TST field.
Multi-organ Segmentation via Co-training Weight-averaged Models from Few-organ Datasets
Aug 17, 2020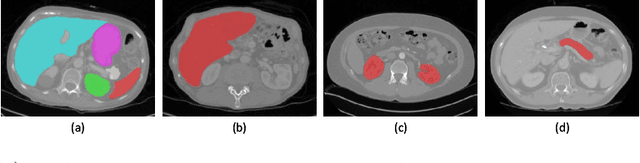
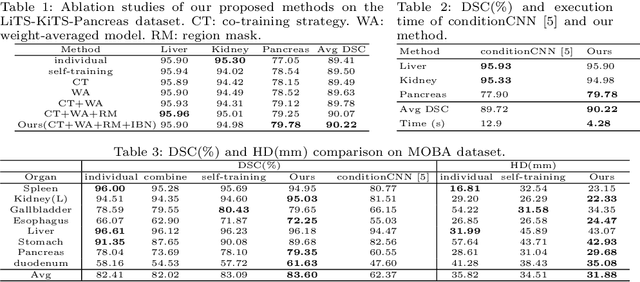
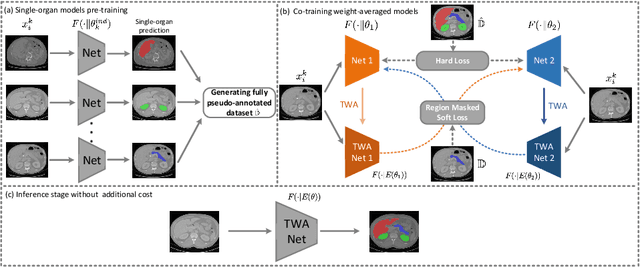
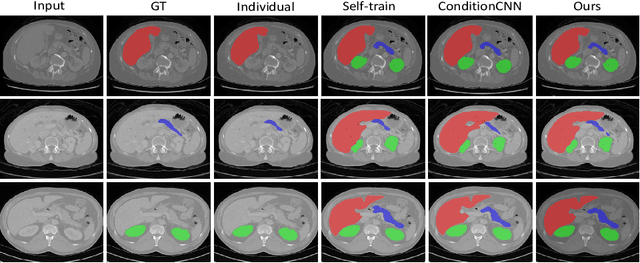
Abstract:Multi-organ segmentation has extensive applications in many clinical applications. To segment multiple organs of interest, it is generally quite difficult to collect full annotations of all the organs on the same images, as some medical centers might only annotate a portion of the organs due to their own clinical practice. In most scenarios, one might obtain annotations of a single or a few organs from one training set, and obtain annotations of the the other organs from another set of training images. Existing approaches mostly train and deploy a single model for each subset of organs, which are memory intensive and also time inefficient. In this paper, we propose to co-train weight-averaged models for learning a unified multi-organ segmentation network from few-organ datasets. We collaboratively train two networks and let the coupled networks teach each other on un-annotated organs. To alleviate the noisy teaching supervisions between the networks, the weighted-averaged models are adopted to produce more reliable soft labels. In addition, a novel region mask is utilized to selectively apply the consistent constraint on the un-annotated organ regions that require collaborative teaching, which further boosts the performance. Extensive experiments on three public available single-organ datasets LiTS, KiTS, Pancreas and manually-constructed single-organ datasets from MOBA show that our method can better utilize the few-organ datasets and achieves superior performance with less inference computational cost.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020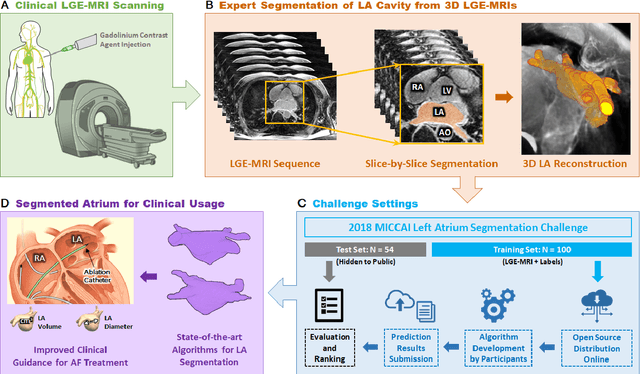
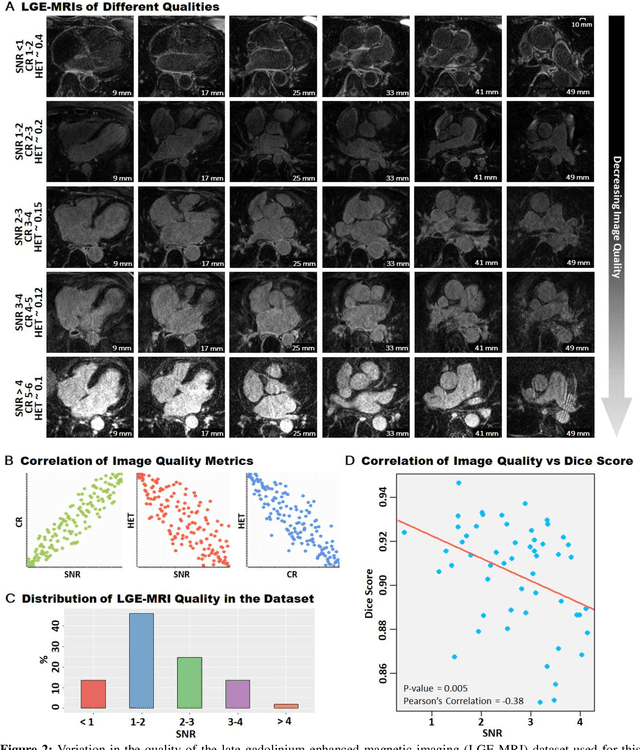
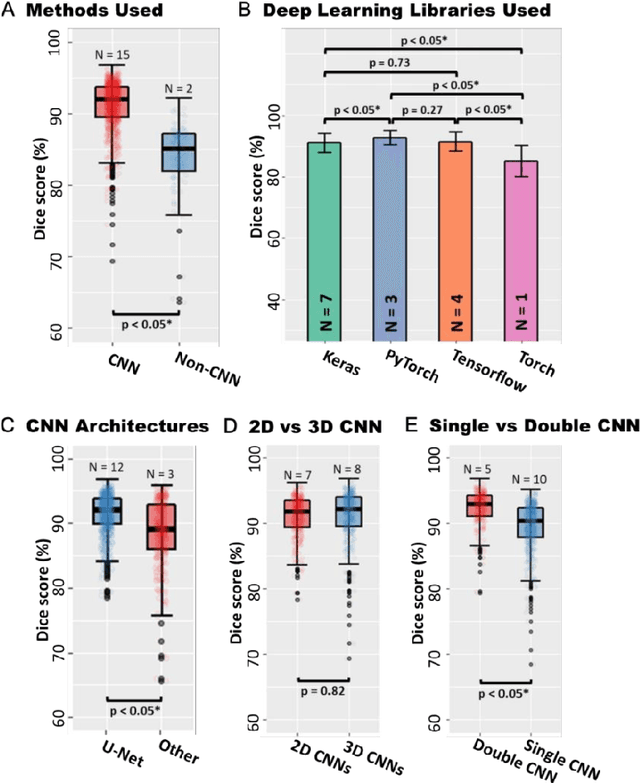
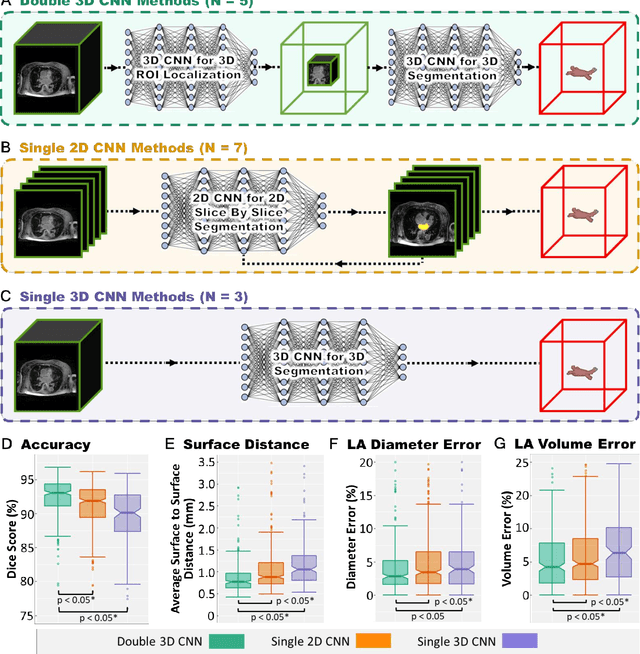
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Large-scale Gastric Cancer Screening and Localization Using Multi-task Deep Neural Network
Oct 12, 2019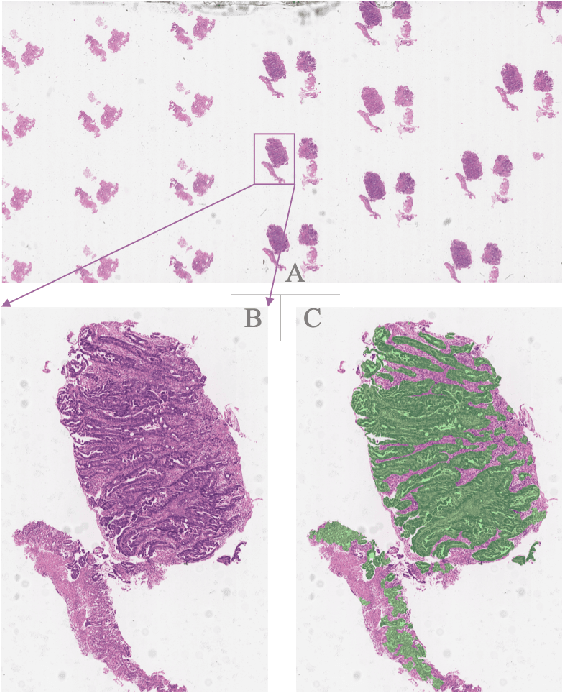
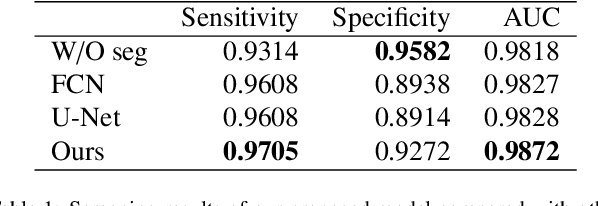
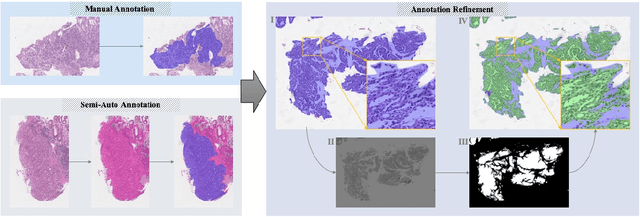
Abstract:Gastric cancer is one of the most common cancers, which ranks third among the leading causes of cancer death. Biopsy of gastric mucosal is a standard procedure in gastric cancer screening test. However, manual pathological inspection is labor-intensive and time-consuming. Besides, it is challenging for an automated algorithm to locate the small lesion regions in the gigapixel whole-slide image and make the decision correctly. To tackle these issues, we collected large-scale whole-slide image dataset with detailed lesion region annotation and designed a whole-slide image analyzing framework consisting of 3 networks which could not only determine the screen result but also present the suspicious areas to the pathologist for reference. Experiments demonstrated that our proposed framework achieves sensitivity of 97.05% and specificity of 92.72% in screening task and Dice coefficient of 0.8331 in segmentation task. Furthermore, we tested our best model in real-world scenario on 10, 316 whole-slide images collected from 4 medical centers.
Accurate Nuclear Segmentation with Center Vector Encoding
Jul 10, 2019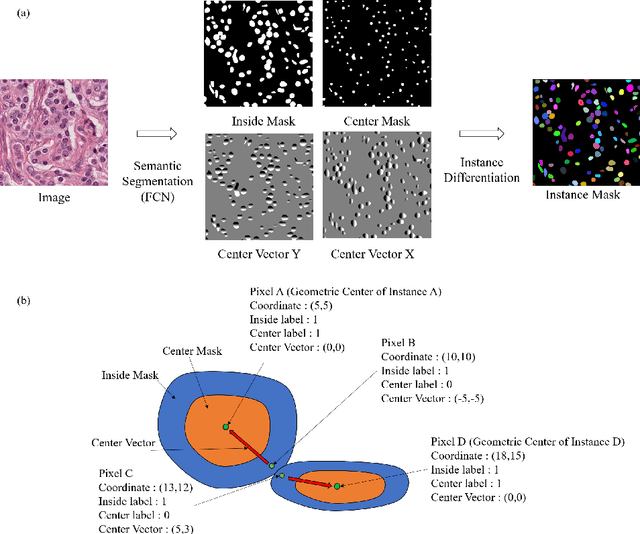
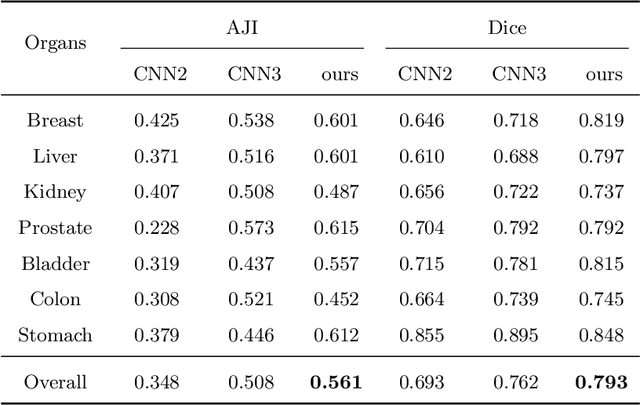
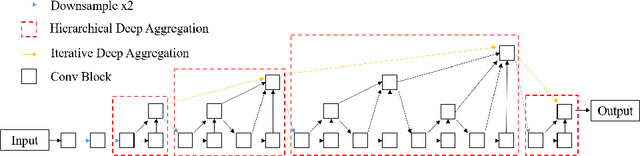
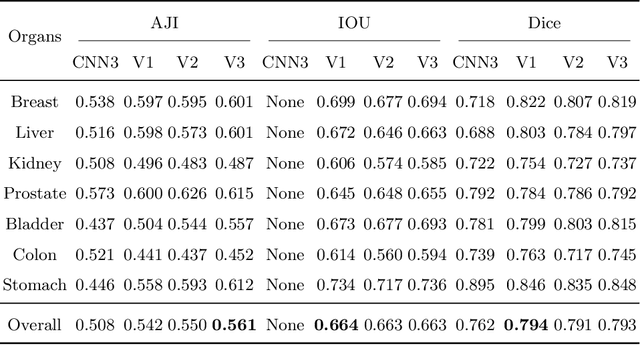
Abstract:Nuclear segmentation is important and frequently demanded for pathology image analysis, yet is also challenging due to nuclear crowdedness and possible occlusion. In this paper, we present a novel bottom-up method for nuclear segmentation. The concepts of Center Mask and Center Vector are introduced to better depict the relationship between pixels and nuclear instances. The instance differentiation process are thus largely simplified and easier to understand. Experiments demonstrate the effectiveness of Center Vector Encoding, where our method outperforms state-of-the-arts by a clear margin.
Signet Ring Cell Detection With a Semi-supervised Learning Framework
Jul 09, 2019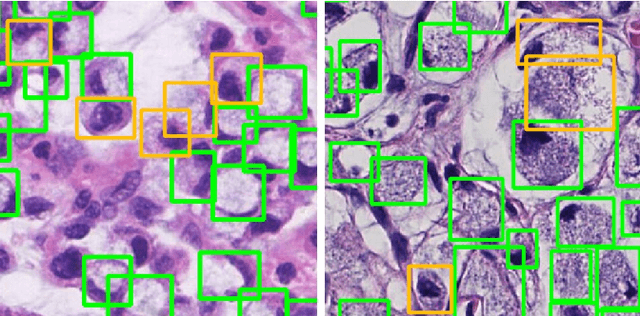
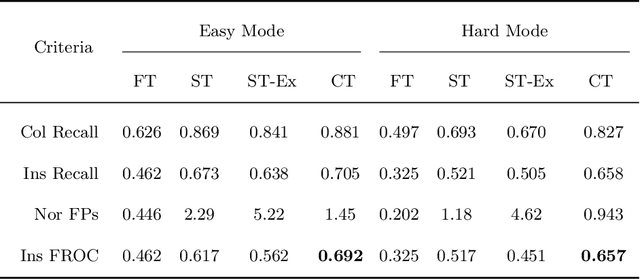
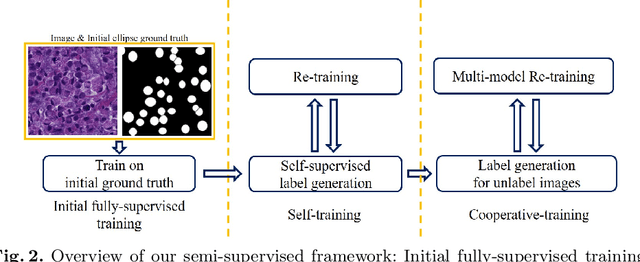
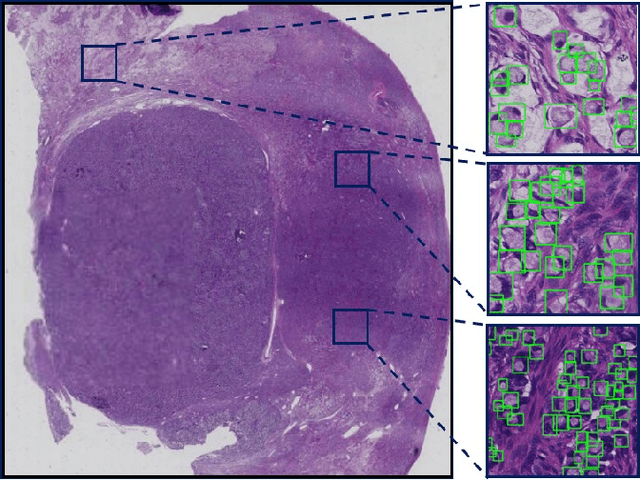
Abstract:Signet ring cell carcinoma is a type of rare adenocarcinoma with poor prognosis. Early detection leads to huge improvement of patients' survival rate. However, pathologists can only visually detect signet ring cells under the microscope. This procedure is not only laborious but also prone to omission. An automatic and accurate signet ring cell detection solution is thus important but has not been investigated before. In this paper, we take the first step to present a semi-supervised learning framework for the signet ring cell detection problem. Self-training is proposed to deal with the challenge of incomplete annotations, and cooperative-training is adapted to explore the unlabeled regions. Combining the two techniques, our semi-supervised learning framework can make better use of both labeled and unlabeled data. Experiments on large real clinical data demonstrate the effectiveness of our design. Our framework achieves accurate signet ring cell detection and can be readily applied in the clinical trails. The dataset will be released soon to facilitate the development of the area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge