Zehao Yu
University of Tuebingen, Tuebingen AI Center, Germany
Identifying Symptoms of Delirium from Clinical Narratives Using Natural Language Processing
Mar 31, 2023



Abstract:Delirium is an acute decline or fluctuation in attention, awareness, or other cognitive function that can lead to serious adverse outcomes. Despite the severe outcomes, delirium is frequently unrecognized and uncoded in patients' electronic health records (EHRs) due to its transient and diverse nature. Natural language processing (NLP), a key technology that extracts medical concepts from clinical narratives, has shown great potential in studies of delirium outcomes and symptoms. To assist in the diagnosis and phenotyping of delirium, we formed an expert panel to categorize diverse delirium symptoms, composed annotation guidelines, created a delirium corpus with diverse delirium symptoms, and developed NLP methods to extract delirium symptoms from clinical notes. We compared 5 state-of-the-art transformer models including 2 models (BERT and RoBERTa) from the general domain and 3 models (BERT_MIMIC, RoBERTa_MIMIC, and GatorTron) from the clinical domain. GatorTron achieved the best strict and lenient F1 scores of 0.8055 and 0.8759, respectively. We conducted an error analysis to identify challenges in annotating delirium symptoms and developing NLP systems. To the best of our knowledge, this is the first large language model-based delirium symptom extraction system. Our study lays the foundation for the future development of computable phenotypes and diagnosis methods for delirium.
Extracting Thyroid Nodules Characteristics from Ultrasound Reports Using Transformer-based Natural Language Processing Methods
Mar 31, 2023



Abstract:The ultrasound characteristics of thyroid nodules guide the evaluation of thyroid cancer in patients with thyroid nodules. However, the characteristics of thyroid nodules are often documented in clinical narratives such as ultrasound reports. Previous studies have examined natural language processing (NLP) methods in extracting a limited number of characteristics (<9) using rule-based NLP systems. In this study, a multidisciplinary team of NLP experts and thyroid specialists, identified thyroid nodule characteristics that are important for clinical care, composed annotation guidelines, developed a corpus, and compared 5 state-of-the-art transformer-based NLP methods, including BERT, RoBERTa, LongFormer, DeBERTa, and GatorTron, for extraction of thyroid nodule characteristics from ultrasound reports. Our GatorTron model, a transformer-based large language model trained using over 90 billion words of text, achieved the best strict and lenient F1-score of 0.8851 and 0.9495 for the extraction of a total number of 16 thyroid nodule characteristics, and 0.9321 for linking characteristics to nodules, outperforming other clinical transformer models. To the best of our knowledge, this is the first study to systematically categorize and apply transformer-based NLP models to extract a large number of clinical relevant thyroid nodule characteristics from ultrasound reports. This study lays ground for assessing the documentation quality of thyroid ultrasound reports and examining outcomes of patients with thyroid nodules using electronic health records.
Contextualized Medication Information Extraction Using Transformer-based Deep Learning Architectures
Mar 14, 2023Abstract:Objective: To develop a natural language processing (NLP) system to extract medications and contextual information that help understand drug changes. This project is part of the 2022 n2c2 challenge. Materials and methods: We developed NLP systems for medication mention extraction, event classification (indicating medication changes discussed or not), and context classification to classify medication changes context into 5 orthogonal dimensions related to drug changes. We explored 6 state-of-the-art pretrained transformer models for the three subtasks, including GatorTron, a large language model pretrained using >90 billion words of text (including >80 billion words from >290 million clinical notes identified at the University of Florida Health). We evaluated our NLP systems using annotated data and evaluation scripts provided by the 2022 n2c2 organizers. Results:Our GatorTron models achieved the best F1-scores of 0.9828 for medication extraction (ranked 3rd), 0.9379 for event classification (ranked 2nd), and the best micro-average accuracy of 0.9126 for context classification. GatorTron outperformed existing transformer models pretrained using smaller general English text and clinical text corpora, indicating the advantage of large language models. Conclusion: This study demonstrated the advantage of using large transformer models for contextual medication information extraction from clinical narratives.
Clinical Concept and Relation Extraction Using Prompt-based Machine Reading Comprehension
Mar 14, 2023



Abstract:Objective: To develop a natural language processing system that solves both clinical concept extraction and relation extraction in a unified prompt-based machine reading comprehension (MRC) architecture with good generalizability for cross-institution applications. Methods: We formulate both clinical concept extraction and relation extraction using a unified prompt-based MRC architecture and explore state-of-the-art transformer models. We compare our MRC models with existing deep learning models for concept extraction and end-to-end relation extraction using two benchmark datasets developed by the 2018 National NLP Clinical Challenges (n2c2) challenge (medications and adverse drug events) and the 2022 n2c2 challenge (relations of social determinants of health [SDoH]). We also evaluate the transfer learning ability of the proposed MRC models in a cross-institution setting. We perform error analyses and examine how different prompting strategies affect the performance of MRC models. Results and Conclusion: The proposed MRC models achieve state-of-the-art performance for clinical concept and relation extraction on the two benchmark datasets, outperforming previous non-MRC transformer models. GatorTron-MRC achieves the best strict and lenient F1-scores for concept extraction, outperforming previous deep learning models on the two datasets by 1%~3% and 0.7%~1.3%, respectively. For end-to-end relation extraction, GatorTron-MRC and BERT-MIMIC-MRC achieve the best F1-scores, outperforming previous deep learning models by 0.9%~2.4% and 10%-11%, respectively. For cross-institution evaluation, GatorTron-MRC outperforms traditional GatorTron by 6.4% and 16% for the two datasets, respectively. The proposed method is better at handling nested/overlapped concepts, extracting relations, and has good portability for cross-institute applications.
SODA: A Natural Language Processing Package to Extract Social Determinants of Health for Cancer Studies
Dec 06, 2022



Abstract:Objective: We aim to develop an open-source natural language processing (NLP) package, SODA (i.e., SOcial DeterminAnts), with pre-trained transformer models to extract social determinants of health (SDoH) for cancer patients, examine the generalizability of SODA to a new disease domain (i.e., opioid use), and evaluate the extraction rate of SDoH using cancer populations. Methods: We identified SDoH categories and attributes and developed an SDoH corpus using clinical notes from a general cancer cohort. We compared four transformer-based NLP models to extract SDoH, examined the generalizability of NLP models to a cohort of patients prescribed with opioids, and explored customization strategies to improve performance. We applied the best NLP model to extract 19 categories of SDoH from the breast (n=7,971), lung (n=11,804), and colorectal cancer (n=6,240) cohorts. Results and Conclusion: We developed a corpus of 629 cancer patients notes with annotations of 13,193 SDoH concepts/attributes from 19 categories of SDoH. The Bidirectional Encoder Representations from Transformers (BERT) model achieved the best strict/lenient F1 scores of 0.9216 and 0.9441 for SDoH concept extraction, 0.9617 and 0.9626 for linking attributes to SDoH concepts. Fine-tuning the NLP models using new annotations from opioid use patients improved the strict/lenient F1 scores from 0.8172/0.8502 to 0.8312/0.8679. The extraction rates among 19 categories of SDoH varied greatly, where 10 SDoH could be extracted from >70% of cancer patients, but 9 SDoH had a low extraction rate (<70% of cancer patients). The SODA package with pre-trained transformer models is publicly available at https://github.com/uf-hobiinformatics-lab/SDoH_SODA.
PlaneDepth: Plane-Based Self-Supervised Monocular Depth Estimation
Oct 04, 2022
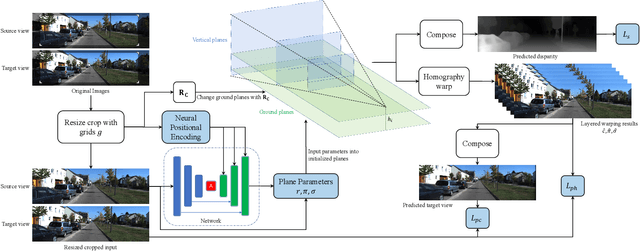


Abstract:Self-supervised monocular depth estimation refers to training a monocular depth estimation (MDE) network using only RGB images to overcome the difficulty of collecting dense ground truth depth. Many previous works addressed this problem using depth classification or depth regression. However, depth classification tends to fall into local minima due to the bilinear interpolation search on the target view. Depth classification overcomes this problem using pre-divided depth bins, but those depth candidates lead to discontinuities in the final depth result, and using the same probability for weighted summation of color and depth is ambiguous. To overcome these limitations, we use some predefined planes that are parallel to the ground, allowing us to automatically segment the ground and predict continuous depth for it. We further model depth as a mixture Laplace distribution, which provides a more certain objective for optimization. Previous works have shown that MDE networks only use the vertical image position of objects to estimate the depth and ignore relative sizes. We address this problem for the first time in both stereo and monocular training using resize cropping data augmentation. Based on our analysis of resize cropping, we combine it with our plane definition and improve our training strategy so that the network could learn the relationship between depth and both the vertical image position and relative size of objects. We further combine the self-distillation stage with post-processing to provide more accurate supervision and save extra time in post-processing. We conduct extensive experiments to demonstrate the effectiveness of our analysis and improvements.
MonoSDF: Exploring Monocular Geometric Cues for Neural Implicit Surface Reconstruction
Jun 01, 2022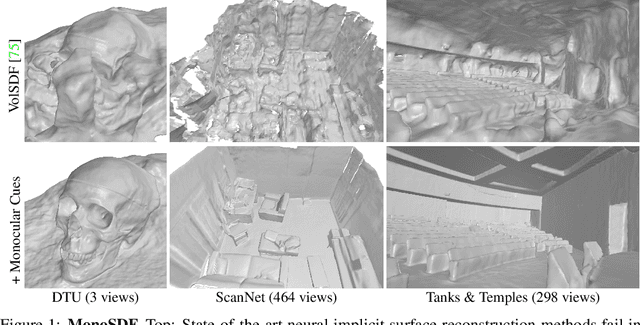
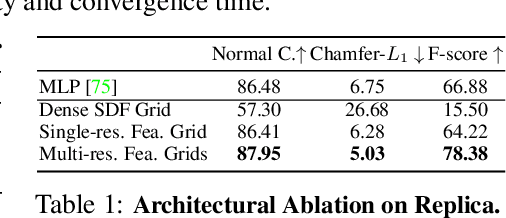
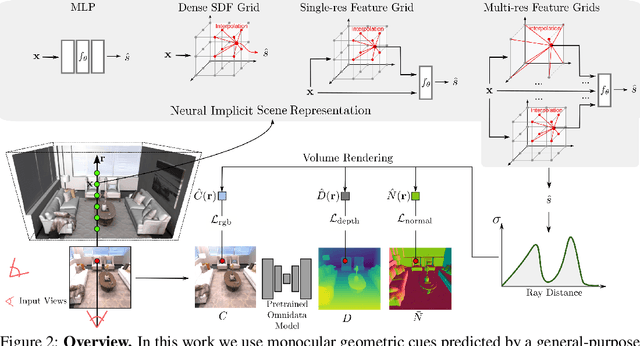
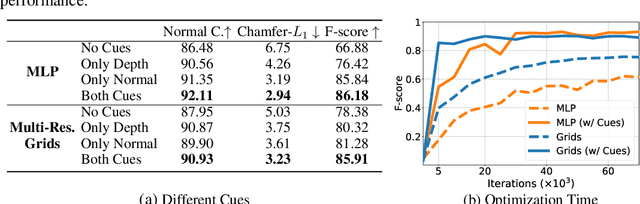
Abstract:In recent years, neural implicit surface reconstruction methods have become popular for multi-view 3D reconstruction. In contrast to traditional multi-view stereo methods, these approaches tend to produce smoother and more complete reconstructions due to the inductive smoothness bias of neural networks. State-of-the-art neural implicit methods allow for high-quality reconstructions of simple scenes from many input views. Yet, their performance drops significantly for larger and more complex scenes and scenes captured from sparse viewpoints. This is caused primarily by the inherent ambiguity in the RGB reconstruction loss that does not provide enough constraints, in particular in less-observed and textureless areas. Motivated by recent advances in the area of monocular geometry prediction, we systematically explore the utility these cues provide for improving neural implicit surface reconstruction. We demonstrate that depth and normal cues, predicted by general-purpose monocular estimators, significantly improve reconstruction quality and optimization time. Further, we analyse and investigate multiple design choices for representing neural implicit surfaces, ranging from monolithic MLP models over single-grid to multi-resolution grid representations. We observe that geometric monocular priors improve performance both for small-scale single-object as well as large-scale multi-object scenes, independent of the choice of representation.
TransFuser: Imitation with Transformer-Based Sensor Fusion for Autonomous Driving
May 31, 2022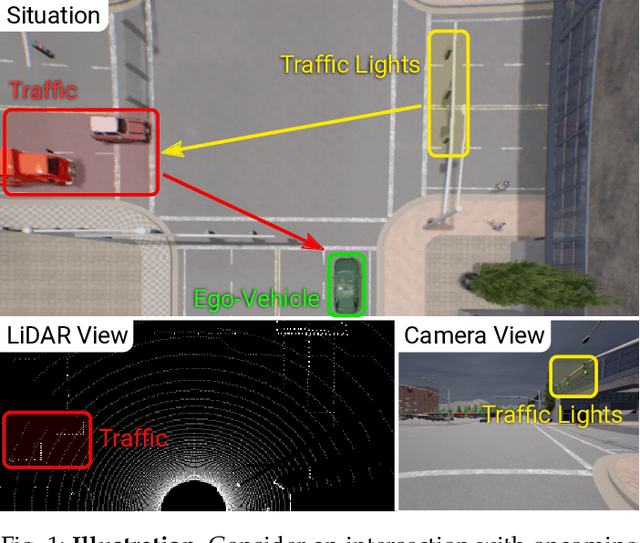
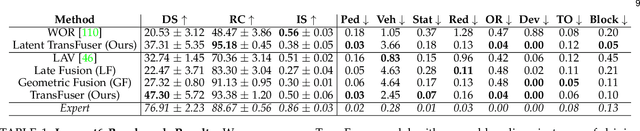
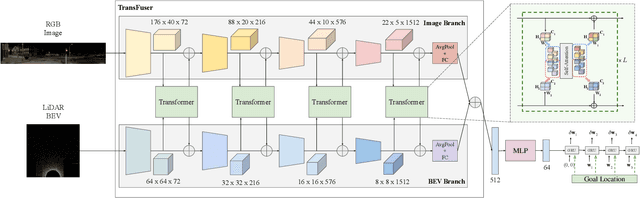
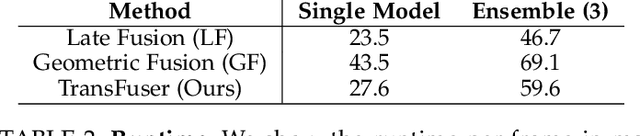
Abstract:How should we integrate representations from complementary sensors for autonomous driving? Geometry-based fusion has shown promise for perception (e.g. object detection, motion forecasting). However, in the context of end-to-end driving, we find that imitation learning based on existing sensor fusion methods underperforms in complex driving scenarios with a high density of dynamic agents. Therefore, we propose TransFuser, a mechanism to integrate image and LiDAR representations using self-attention. Our approach uses transformer modules at multiple resolutions to fuse perspective view and bird's eye view feature maps. We experimentally validate its efficacy on a challenging new benchmark with long routes and dense traffic, as well as the official leaderboard of the CARLA urban driving simulator. At the time of submission, TransFuser outperforms all prior work on the CARLA leaderboard in terms of driving score by a large margin. Compared to geometry-based fusion, TransFuser reduces the average collisions per kilometer by 48%.
Clinical Relation Extraction Using Transformer-based Models
Aug 16, 2021
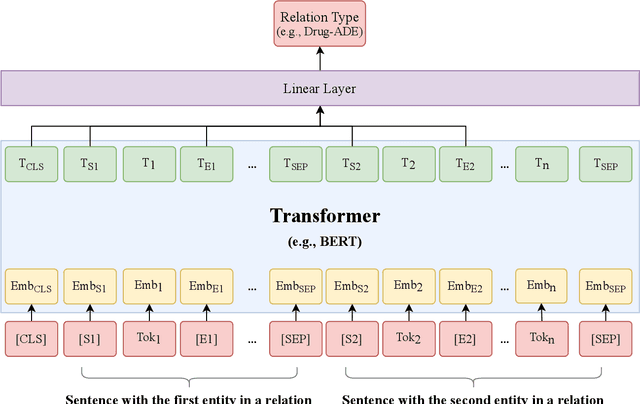
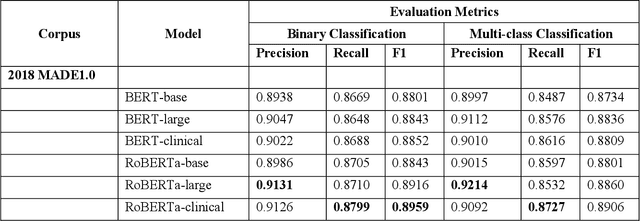
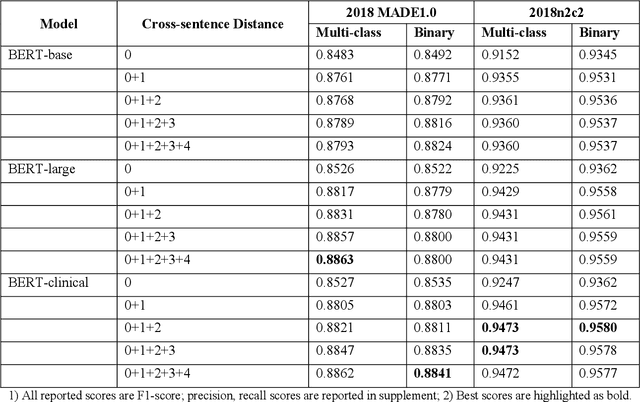
Abstract:The newly emerged transformer technology has a tremendous impact on NLP research. In the general English domain, transformer-based models have achieved state-of-the-art performances on various NLP benchmarks. In the clinical domain, researchers also have investigated transformer models for clinical applications. The goal of this study is to systematically explore three widely used transformer-based models (i.e., BERT, RoBERTa, and XLNet) for clinical relation extraction and develop an open-source package with clinical pre-trained transformer-based models to facilitate information extraction in the clinical domain. We developed a series of clinical RE models based on three transformer architectures, namely BERT, RoBERTa, and XLNet. We evaluated these models using 2 publicly available datasets from 2018 MADE1.0 and 2018 n2c2 challenges. We compared two classification strategies (binary vs. multi-class classification) and investigated two approaches to generate candidate relations in different experimental settings. In this study, we compared three transformer-based (BERT, RoBERTa, and XLNet) models for relation extraction. We demonstrated that the RoBERTa-clinical RE model achieved the best performance on the 2018 MADE1.0 dataset with an F1-score of 0.8958. On the 2018 n2c2 dataset, the XLNet-clinical model achieved the best F1-score of 0.9610. Our results indicated that the binary classification strategy consistently outperformed the multi-class classification strategy for clinical relation extraction. Our methods and models are publicly available at https://github.com/uf-hobi-informatics-lab/ClinicalTransformerRelationExtraction. We believe this work will improve current practice on clinical relation extraction and other related NLP tasks in the biomedical domain.
A Study of Social and Behavioral Determinants of Health in Lung Cancer Patients Using Transformers-based Natural Language Processing Models
Aug 10, 2021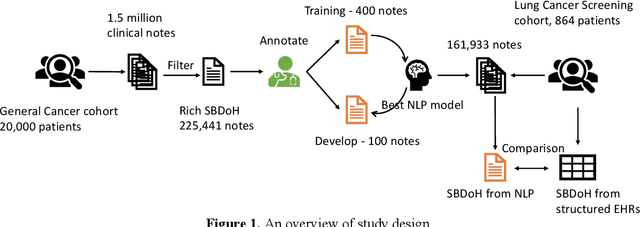
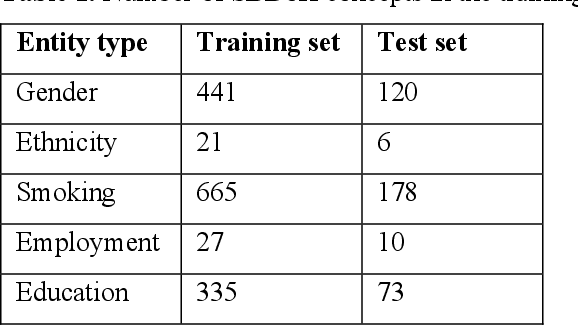
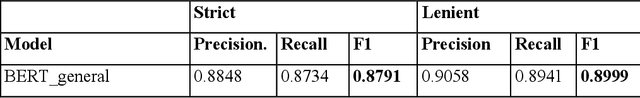
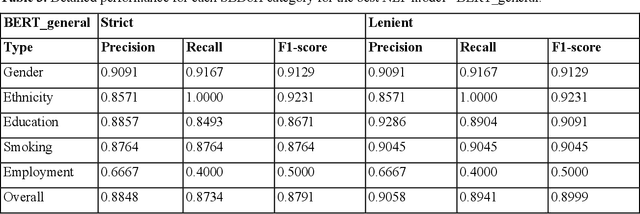
Abstract:Social and behavioral determinants of health (SBDoH) have important roles in shaping people's health. In clinical research studies, especially comparative effectiveness studies, failure to adjust for SBDoH factors will potentially cause confounding issues and misclassification errors in either statistical analyses and machine learning-based models. However, there are limited studies to examine SBDoH factors in clinical outcomes due to the lack of structured SBDoH information in current electronic health record (EHR) systems, while much of the SBDoH information is documented in clinical narratives. Natural language processing (NLP) is thus the key technology to extract such information from unstructured clinical text. However, there is not a mature clinical NLP system focusing on SBDoH. In this study, we examined two state-of-the-art transformer-based NLP models, including BERT and RoBERTa, to extract SBDoH concepts from clinical narratives, applied the best performing model to extract SBDoH concepts on a lung cancer screening patient cohort, and examined the difference of SBDoH information between NLP extracted results and structured EHRs (SBDoH information captured in standard vocabularies such as the International Classification of Diseases codes). The experimental results show that the BERT-based NLP model achieved the best strict/lenient F1-score of 0.8791 and 0.8999, respectively. The comparison between NLP extracted SBDoH information and structured EHRs in the lung cancer patient cohort of 864 patients with 161,933 various types of clinical notes showed that much more detailed information about smoking, education, and employment were only captured in clinical narratives and that it is necessary to use both clinical narratives and structured EHRs to construct a more complete picture of patients' SBDoH factors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge