Yvonne W. Lui
Pixelwise Uncertainty Quantification of Accelerated MRI Reconstruction
Jan 19, 2026Abstract:Parallel imaging techniques reduce magnetic resonance imaging (MRI) scan time but image quality degrades as the acceleration factor increases. In clinical practice, conservative acceleration factors are chosen because no mechanism exists to automatically assess the diagnostic quality of undersampled reconstructions. This work introduces a general framework for pixel-wise uncertainty quantification in parallel MRI reconstructions, enabling automatic identification of unreliable regions without access to any ground-truth reference image. Our method integrates conformal quantile regression with image reconstruction methods to estimate statistically rigorous pixel-wise uncertainty intervals. We trained and evaluated our model on Cartesian undersampled brain and knee data obtained from the fastMRI dataset using acceleration factors ranging from 2 to 10. An end-to-end Variational Network was used for image reconstruction. Quantitative experiments demonstrate strong agreement between predicted uncertainty maps and true reconstruction error. Using our method, the corresponding Pearson correlation coefficient was higher than 90% at acceleration levels at and above four-fold; whereas it dropped to less than 70% when the uncertainty was computed using a simpler a heuristic notion (magnitude of the residual). Qualitative examples further show the uncertainty maps based on quantile regression capture the magnitude and spatial distribution of reconstruction errors across acceleration factors, with regions of elevated uncertainty aligning with pathologies and artifacts. The proposed framework enables evaluation of reconstruction quality without access to fully-sampled ground-truth reference images. It represents a step toward adaptive MRI acquisition protocols that may be able to dynamically balance scan time and diagnostic reliability.
A Non-contrast Head CT Foundation Model for Comprehensive Neuro-Trauma Triage
Feb 28, 2025



Abstract:Recent advancements in AI and medical imaging offer transformative potential in emergency head CT interpretation for reducing assessment times and improving accuracy in the face of an increasing request of such scans and a global shortage in radiologists. This study introduces a 3D foundation model for detecting diverse neuro-trauma findings with high accuracy and efficiency. Using large language models (LLMs) for automatic labeling, we generated comprehensive multi-label annotations for critical conditions. Our approach involved pretraining neural networks for hemorrhage subtype segmentation and brain anatomy parcellation, which were integrated into a pretrained comprehensive neuro-trauma detection network through multimodal fine-tuning. Performance evaluation against expert annotations and comparison with CT-CLIP demonstrated strong triage accuracy across major neuro-trauma findings, such as hemorrhage and midline shift, as well as less frequent critical conditions such as cerebral edema and arterial hyperdensity. The integration of neuro-specific features significantly enhanced diagnostic capabilities, achieving an average AUC of 0.861 for 16 neuro-trauma conditions. This work advances foundation models in medical imaging, serving as a benchmark for future AI-assisted neuro-trauma diagnostics in emergency radiology.
fastMRI Breast: A publicly available radial k-space dataset of breast dynamic contrast-enhanced MRI
Jun 07, 2024

Abstract:This data curation work introduces the first large-scale dataset of radial k-space and DICOM data for breast DCE-MRI acquired in diagnostic breast MRI exams. Our dataset includes case-level labels indicating patient age, menopause status, lesion status (negative, benign, and malignant), and lesion type for each case. The public availability of this dataset and accompanying reconstruction code will support research and development of fast and quantitative breast image reconstruction and machine learning methods.
fastMRI+: Clinical Pathology Annotations for Knee and Brain Fully Sampled Multi-Coil MRI Data
Sep 14, 2021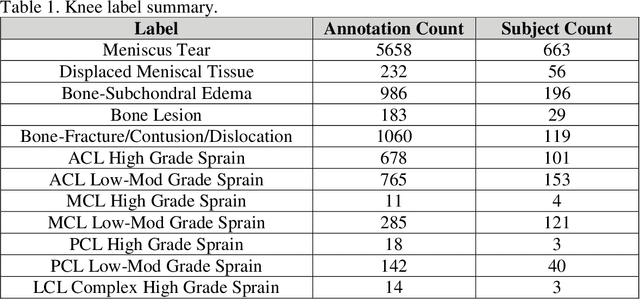
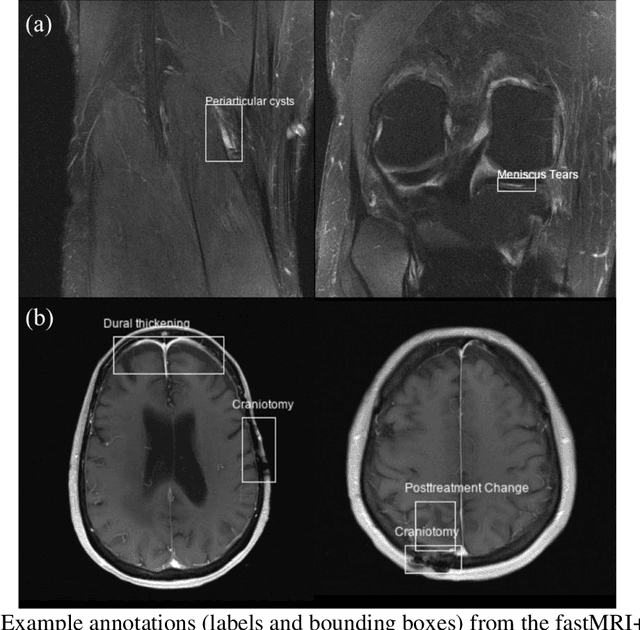
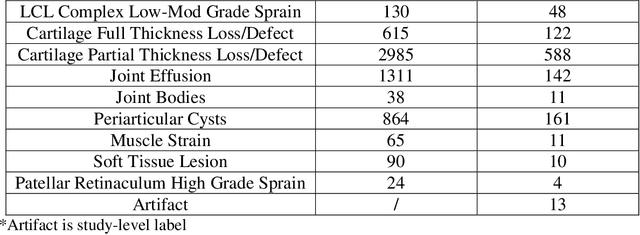
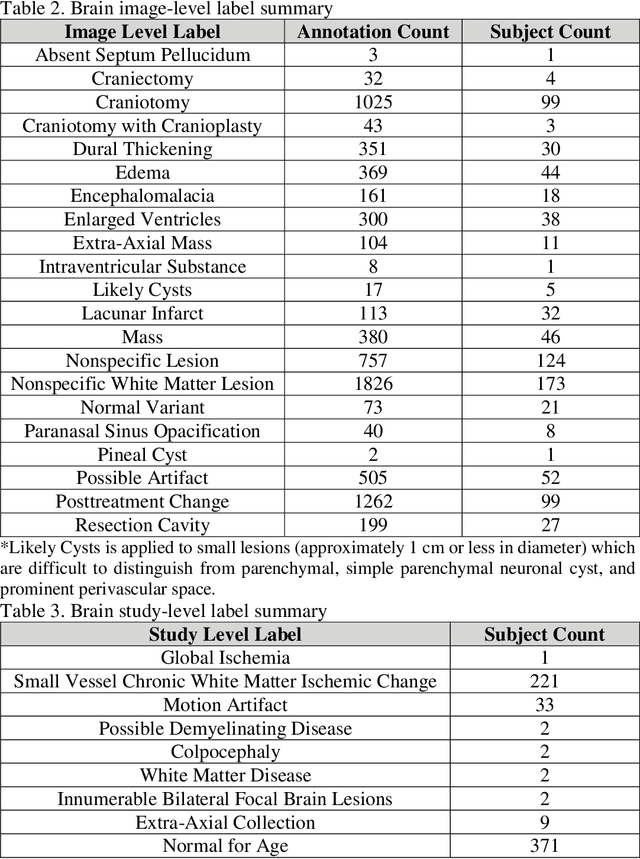
Abstract:Improving speed and image quality of Magnetic Resonance Imaging (MRI) via novel reconstruction approaches remains one of the highest impact applications for deep learning in medical imaging. The fastMRI dataset, unique in that it contains large volumes of raw MRI data, has enabled significant advances in accelerating MRI using deep learning-based reconstruction methods. While the impact of the fastMRI dataset on the field of medical imaging is unquestioned, the dataset currently lacks clinical expert pathology annotations, critical to addressing clinically relevant reconstruction frameworks and exploring important questions regarding rendering of specific pathology using such novel approaches. This work introduces fastMRI+, which consists of 16154 subspecialist expert bounding box annotations and 13 study-level labels for 22 different pathology categories on the fastMRI knee dataset, and 7570 subspecialist expert bounding box annotations and 643 study-level labels for 30 different pathology categories for the fastMRI brain dataset. The fastMRI+ dataset is open access and aims to support further research and advancement of medical imaging in MRI reconstruction and beyond.
An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department
Aug 04, 2020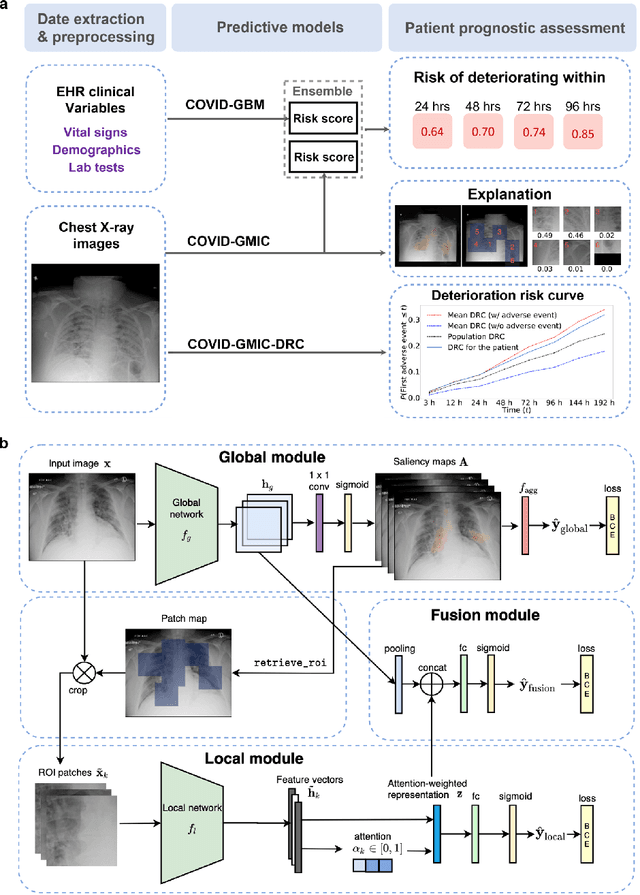
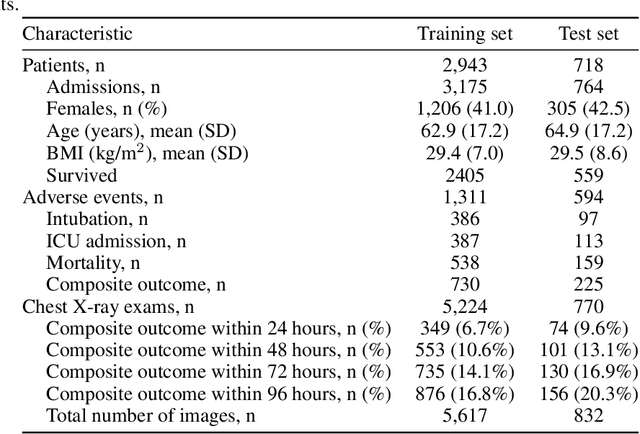
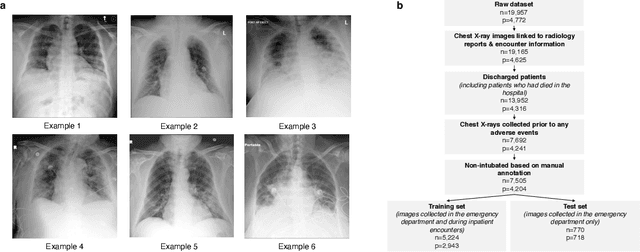
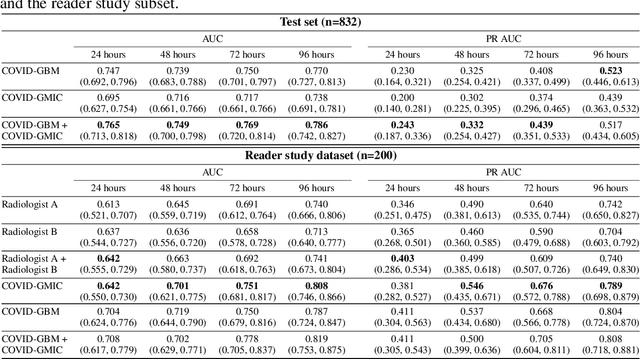
Abstract:During the COVID-19 pandemic, rapid and accurate triage of patients at the emergency department is critical to inform decision-making. We propose a data-driven approach for automatic prediction of deterioration risk using a deep neural network that learns from chest X-ray images, and a gradient boosting model that learns from routine clinical variables. Our AI prognosis system, trained using data from 3,661 patients, achieves an AUC of 0.786 (95% CI: 0.742-0.827) when predicting deterioration within 96 hours. The deep neural network extracts informative areas of chest X-ray images to assist clinicians in interpreting the predictions, and performs comparably to two radiologists in a reader study. In order to verify performance in a real clinical setting, we silently deployed a preliminary version of the deep neural network at NYU Langone Health during the first wave of the pandemic, which produced accurate predictions in real-time. In summary, our findings demonstrate the potential of the proposed system for assisting front-line physicians in the triage of COVID-19 patients.
DARTS: DenseUnet-based Automatic Rapid Tool for brain Segmentation
Nov 14, 2019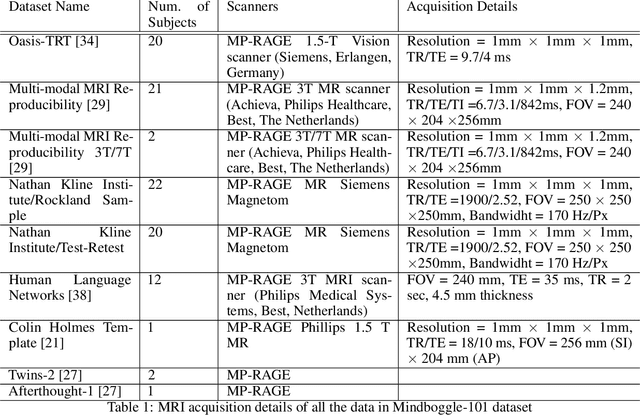
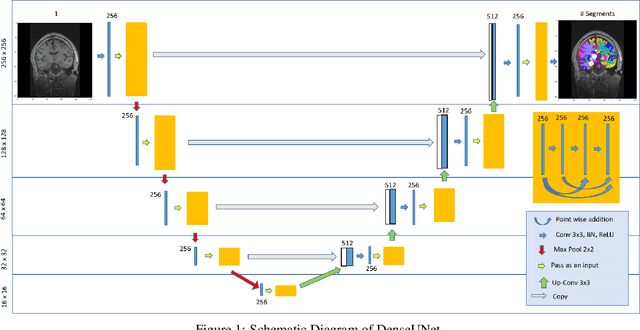
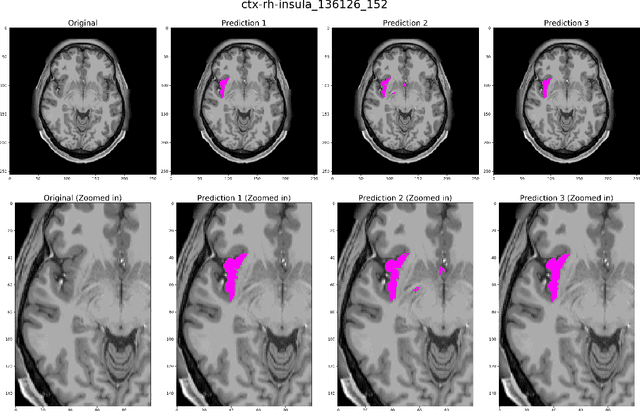
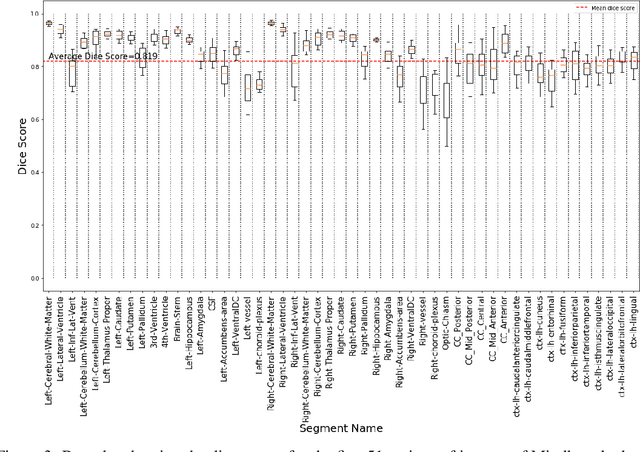
Abstract:Quantitative, volumetric analysis of Magnetic Resonance Imaging (MRI) is a fundamental way researchers study the brain in a host of neurological conditions including normal maturation and aging. Despite the availability of open-source brain segmentation software, widespread clinical adoption of volumetric analysis has been hindered due to processing times and reliance on manual corrections. Here, we extend the use of deep learning models from proof-of-concept, as previously reported, to present a comprehensive segmentation of cortical and deep gray matter brain structures matching the standard regions of aseg+aparc included in the commonly used open-source tool, Freesurfer. The work presented here provides a real-life, rapid deep learning-based brain segmentation tool to enable clinical translation as well as research application of quantitative brain segmentation. The advantages of the presented tool include short (~1 minute) processing time and improved segmentation quality. This is the first study to perform quick and accurate segmentation of 102 brain regions based on the surface-based protocol (DMK protocol), widely used by experts in the field. This is also the first work to include an expert reader study to assess the quality of the segmentation obtained using a deep-learning-based model. We show the superior performance of our deep-learning-based models over the traditional segmentation tool, Freesurfer. We refer to the proposed deep learning-based tool as DARTS (DenseUnet-based Automatic Rapid Tool for brain Segmentation). Our tool and trained models are available at https://github.com/NYUMedML/DARTS
fastMRI: An Open Dataset and Benchmarks for Accelerated MRI
Nov 21, 2018
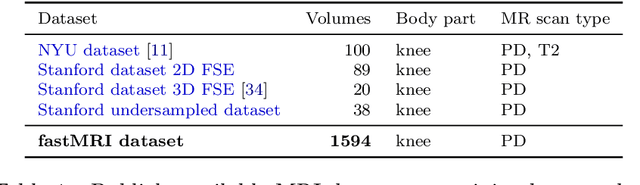
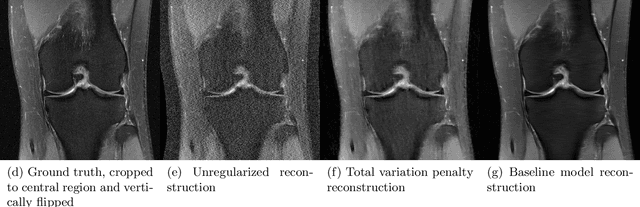
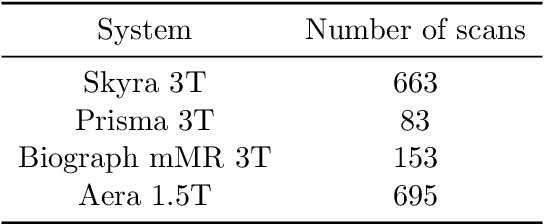
Abstract:Accelerating Magnetic Resonance Imaging (MRI) by taking fewer measurements has the potential to reduce medical costs, minimize stress to patients and make MRI possible in applications where it is currently prohibitively slow or expensive. We introduce the fastMRI dataset, a large-scale collection of both raw MR measurements and clinical MR images, that can be used for training and evaluation of machine-learning approaches to MR image reconstruction. By introducing standardized evaluation criteria and a freely-accessible dataset, our goal is to help the community make rapid advances in the state of the art for MR image reconstruction. We also provide a self-contained introduction to MRI for machine learning researchers with no medical imaging background.
MTBI Identification From Diffusion MR Images Using Bag of Adversarial Visual Features
Jun 27, 2018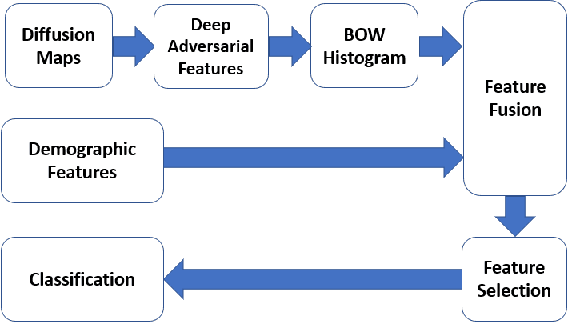
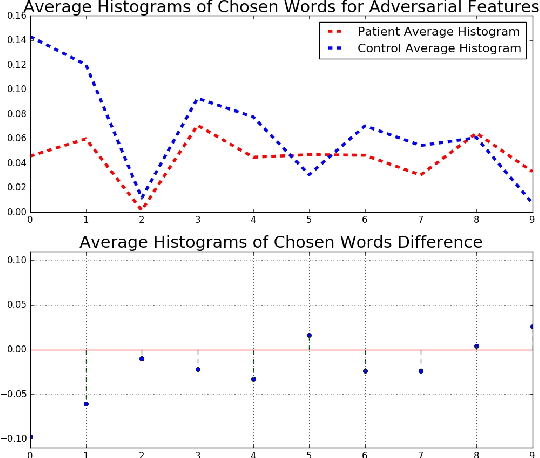
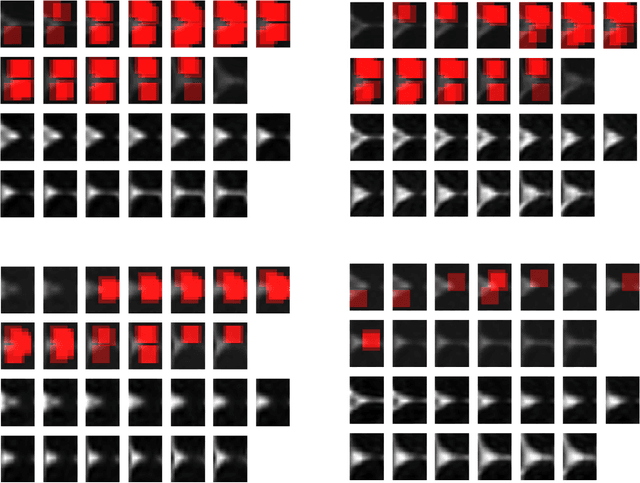
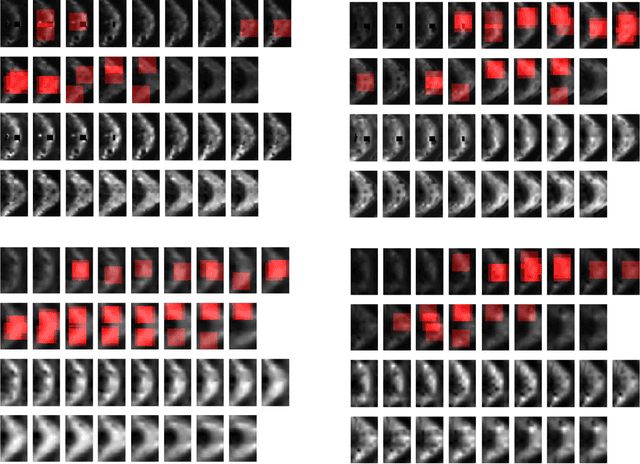
Abstract:In this work, we propose bag of adversarial features (BAF) for identifying mild traumatic brain injury (MTBI) patients from their diffusion magnetic resonance images (MRI) (obtained within one month of injury) by incorporating unsupervised feature learning techniques. MTBI is a growing public health problem with an estimated incidence of over 1.7 million people annually in US. Diagnosis is based on clinical history and symptoms, and accurate, concrete measures of injury are lacking. Unlike most of previous works, which use hand-crafted features extracted from different parts of brain for MTBI classification, we employ feature learning algorithms to learn more discriminative representation for this task. A major challenge in this field thus far is the relatively small number of subjects available for training. This makes it difficult to use an end-to-end convolutional neural network to directly classify a subject from MR images. To overcome this challenge, we first apply an adversarial auto-encoder (with convolutional structure) to learn patch-level features, from overlapping image patches extracted from different brain regions. We then aggregate these features through a bag-of-word approach. We perform an extensive experimental study on a dataset of 227 subjects (including 109 MTBI patients, and 118 age and sex matched healthy controls), and compare the bag-of-deep-features with several previous approaches. Our experimental results show that the BAF significantly outperforms earlier works relying on the mean values of MR metrics in selected brain regions.
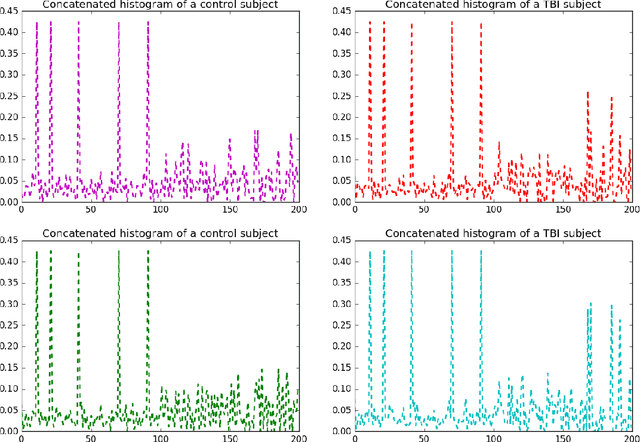
Identifying Mild Traumatic Brain Injury Patients From MR Images Using Bag of Visual Words
Feb 14, 2018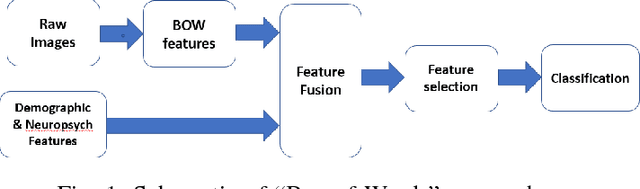
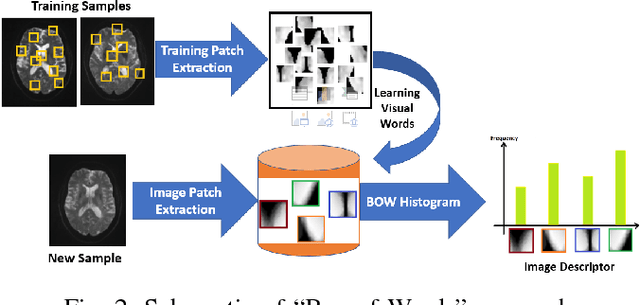
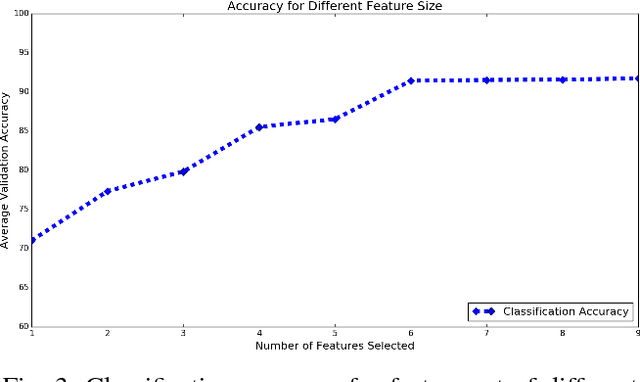
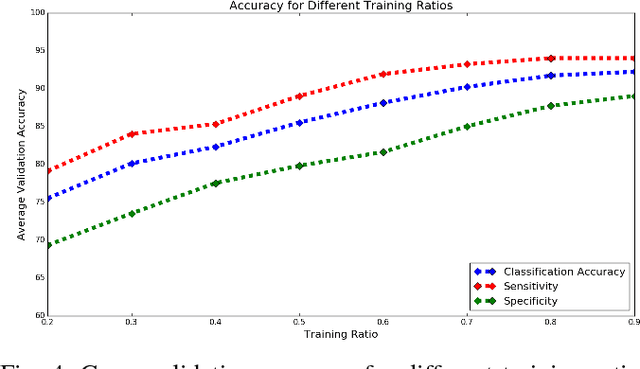
Abstract:Mild traumatic brain injury (mTBI) is a growing public health problem with an estimated incidence of one million people annually in US. Neurocognitive tests are used to both assess the patient condition and to monitor the patient progress. This work aims to directly use MR images taken shortly after injury to detect whether a patient suffers from mTBI, by incorporating machine learning and computer vision techniques to learn features suitable discriminating between mTBI and normal patients. We focus on 3 regions in brain, and extract multiple patches from them, and use bag-of-visual-word technique to represent each subject as a histogram of representative patterns derived from patches from all training subjects. After extracting the features, we use greedy forward feature selection, to choose a subset of features which achieves highest accuracy. We show through experimental studies that BoW features perform better than the simple mean value features which were used previously.
A Machine Learning Approach For Identifying Patients with Mild Traumatic Brain Injury Using Diffusion MRI Modeling
Aug 27, 2017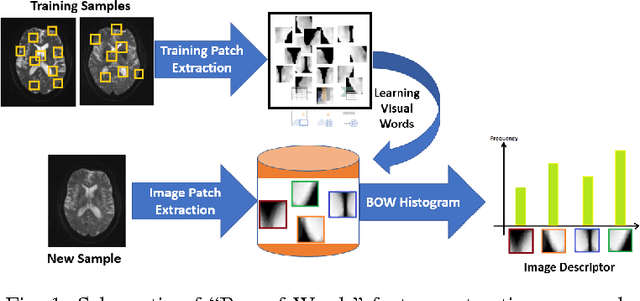
Abstract:While diffusion MRI has been extremely promising in the study of MTBI, identifying patients with recent MTBI remains a challenge. The literature is mixed with regard to localizing injury in these patients, however, gray matter such as the thalamus and white matter including the corpus callosum and frontal deep white matter have been repeatedly implicated as areas at high risk for injury. The purpose of this study is to develop a machine learning framework to classify MTBI patients and controls using features derived from multi-shell diffusion MRI in the thalamus, frontal white matter and corpus callosum.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge