Sasa Grbic
Revisiting 2D Foundation Models for Scalable 3D Medical Image Classification
Dec 15, 2025



Abstract:3D medical image classification is essential for modern clinical workflows. Medical foundation models (FMs) have emerged as a promising approach for scaling to new tasks, yet current research suffers from three critical pitfalls: data-regime bias, suboptimal adaptation, and insufficient task coverage. In this paper, we address these pitfalls and introduce AnyMC3D, a scalable 3D classifier adapted from 2D FMs. Our method scales efficiently to new tasks by adding only lightweight plugins (about 1M parameters per task) on top of a single frozen backbone. This versatile framework also supports multi-view inputs, auxiliary pixel-level supervision, and interpretable heatmap generation. We establish a comprehensive benchmark of 12 tasks covering diverse pathologies, anatomies, and modalities, and systematically analyze state-of-the-art 3D classification techniques. Our analysis reveals key insights: (1) effective adaptation is essential to unlock FM potential, (2) general-purpose FMs can match medical-specific FMs if properly adapted, and (3) 2D-based methods surpass 3D architectures for 3D classification. For the first time, we demonstrate the feasibility of achieving state-of-the-art performance across diverse applications using a single scalable framework (including 1st place in the VLM3D challenge), eliminating the need for separate task-specific models.
AI-assisted Early Detection of Pancreatic Ductal Adenocarcinoma on Contrast-enhanced CT
Mar 13, 2025



Abstract:Pancreatic ductal adenocarcinoma (PDAC) is one of the most common and aggressive types of pancreatic cancer. However, due to the lack of early and disease-specific symptoms, most patients with PDAC are diagnosed at an advanced disease stage. Consequently, early PDAC detection is crucial for improving patients' quality of life and expanding treatment options. In this work, we develop a coarse-to-fine approach to detect PDAC on contrast-enhanced CT scans. First, we localize and crop the region of interest from the low-resolution images, and then segment the PDAC-related structures at a finer scale. Additionally, we introduce two strategies to further boost detection performance: (1) a data-splitting strategy for model ensembling, and (2) a customized post-processing function. We participated in the PANORAMA challenge and ranked 1st place for PDAC detection with an AUROC of 0.9263 and an AP of 0.7243. Our code and models are publicly available at https://github.com/han-liu/PDAC_detection.
A Non-contrast Head CT Foundation Model for Comprehensive Neuro-Trauma Triage
Feb 28, 2025



Abstract:Recent advancements in AI and medical imaging offer transformative potential in emergency head CT interpretation for reducing assessment times and improving accuracy in the face of an increasing request of such scans and a global shortage in radiologists. This study introduces a 3D foundation model for detecting diverse neuro-trauma findings with high accuracy and efficiency. Using large language models (LLMs) for automatic labeling, we generated comprehensive multi-label annotations for critical conditions. Our approach involved pretraining neural networks for hemorrhage subtype segmentation and brain anatomy parcellation, which were integrated into a pretrained comprehensive neuro-trauma detection network through multimodal fine-tuning. Performance evaluation against expert annotations and comparison with CT-CLIP demonstrated strong triage accuracy across major neuro-trauma findings, such as hemorrhage and midline shift, as well as less frequent critical conditions such as cerebral edema and arterial hyperdensity. The integration of neuro-specific features significantly enhanced diagnostic capabilities, achieving an average AUC of 0.861 for 16 neuro-trauma conditions. This work advances foundation models in medical imaging, serving as a benchmark for future AI-assisted neuro-trauma diagnostics in emergency radiology.
Generation of Radiology Findings in Chest X-Ray by Leveraging Collaborative Knowledge
Jun 18, 2023



Abstract:Among all the sub-sections in a typical radiology report, the Clinical Indications, Findings, and Impression often reflect important details about the health status of a patient. The information included in Impression is also often covered in Findings. While Findings and Impression can be deduced by inspecting the image, Clinical Indications often require additional context. The cognitive task of interpreting medical images remains the most critical and often time-consuming step in the radiology workflow. Instead of generating an end-to-end radiology report, in this paper, we focus on generating the Findings from automated interpretation of medical images, specifically chest X-rays (CXRs). Thus, this work focuses on reducing the workload of radiologists who spend most of their time either writing or narrating the Findings. Unlike past research, which addresses radiology report generation as a single-step image captioning task, we have further taken into consideration the complexity of interpreting CXR images and propose a two-step approach: (a) detecting the regions with abnormalities in the image, and (b) generating relevant text for regions with abnormalities by employing a generative large language model (LLM). This two-step approach introduces a layer of interpretability and aligns the framework with the systematic reasoning that radiologists use when reviewing a CXR.
* Information Technology and Quantitative Management (ITQM 2023)
COSST: Multi-organ Segmentation with Partially Labeled Datasets Using Comprehensive Supervisions and Self-training
Apr 28, 2023Abstract:Deep learning models have demonstrated remarkable success in multi-organ segmentation but typically require large-scale datasets with all organs of interest annotated. However, medical image datasets are often low in sample size and only partially labeled, i.e., only a subset of organs are annotated. Therefore, it is crucial to investigate how to learn a unified model on the available partially labeled datasets to leverage their synergistic potential. In this paper, we empirically and systematically study the partial-label segmentation with in-depth analyses on the existing approaches and identify three distinct types of supervision signals, including two signals derived from ground truth and one from pseudo label. We propose a novel training framework termed COSST, which effectively and efficiently integrates comprehensive supervision signals with self-training. Concretely, we first train an initial unified model using two ground truth-based signals and then iteratively incorporate the pseudo label signal to the initial model using self-training. To mitigate performance degradation caused by unreliable pseudo labels, we assess the reliability of pseudo labels via outlier detection in latent space and exclude the most unreliable pseudo labels from each self-training iteration. Extensive experiments are conducted on six CT datasets for three partial-label segmentation tasks. Experimental results show that our proposed COSST achieves significant improvement over the baseline method, i.e., individual networks trained on each partially labeled dataset. Compared to the state-of-the-art partial-label segmentation methods, COSST demonstrates consistent superior performance on various segmentation tasks and with different training data size.
Self-supervised Learning from 100 Million Medical Images
Jan 04, 2022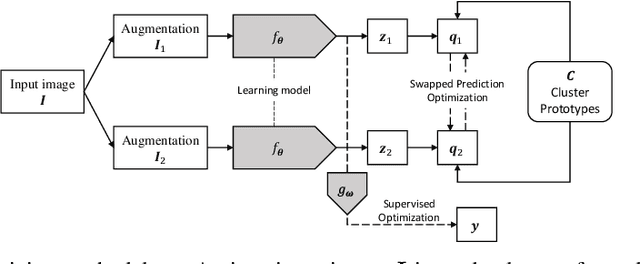
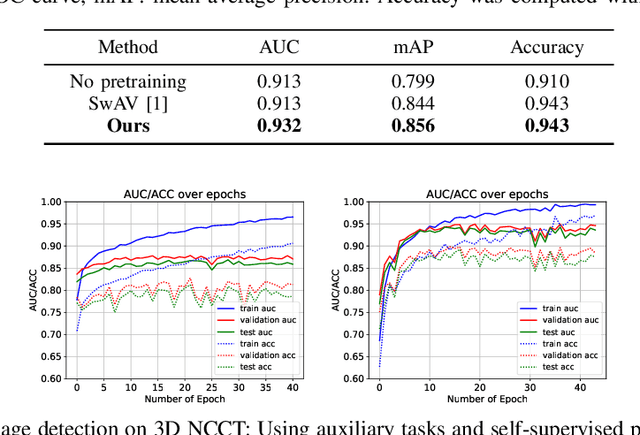
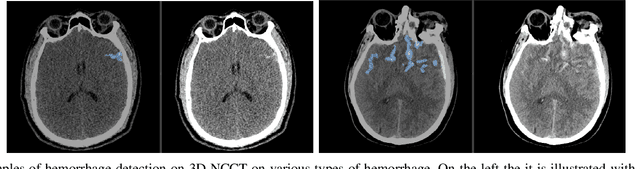
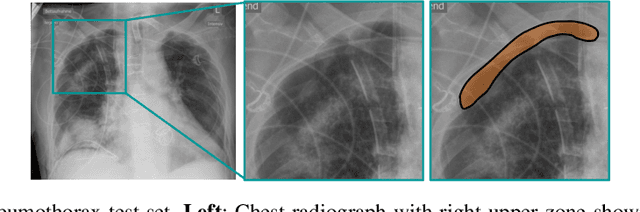
Abstract:Building accurate and robust artificial intelligence systems for medical image assessment requires not only the research and design of advanced deep learning models but also the creation of large and curated sets of annotated training examples. Constructing such datasets, however, is often very costly -- due to the complex nature of annotation tasks and the high level of expertise required for the interpretation of medical images (e.g., expert radiologists). To counter this limitation, we propose a method for self-supervised learning of rich image features based on contrastive learning and online feature clustering. For this purpose we leverage large training datasets of over 100,000,000 medical images of various modalities, including radiography, computed tomography (CT), magnetic resonance (MR) imaging and ultrasonography. We propose to use these features to guide model training in supervised and hybrid self-supervised/supervised regime on various downstream tasks. We highlight a number of advantages of this strategy on challenging image assessment problems in radiography, CT and MR: 1) Significant increase in accuracy compared to the state-of-the-art (e.g., AUC boost of 3-7% for detection of abnormalities from chest radiography scans and hemorrhage detection on brain CT); 2) Acceleration of model convergence during training by up to 85% compared to using no pretraining (e.g., 83% when training a model for detection of brain metastases in MR scans); 3) Increase in robustness to various image augmentations, such as intensity variations, rotations or scaling reflective of data variation seen in the field.
Robust Classification from Noisy Labels: Integrating Additional Knowledge for Chest Radiography Abnormality Assessment
Apr 21, 2021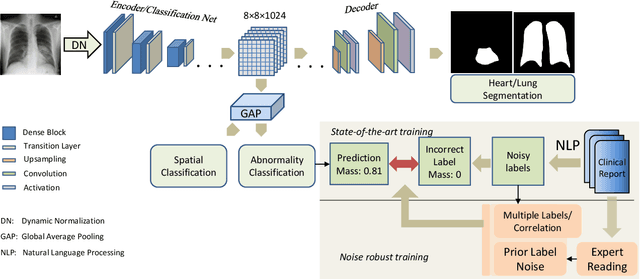
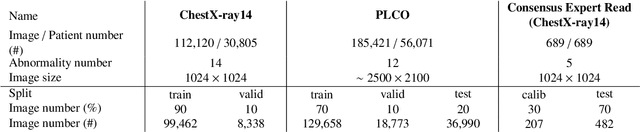
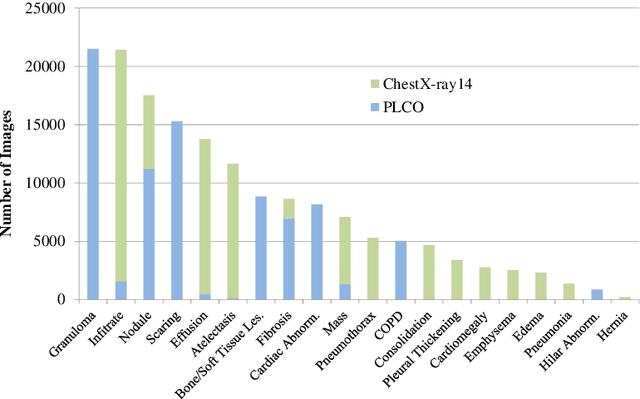
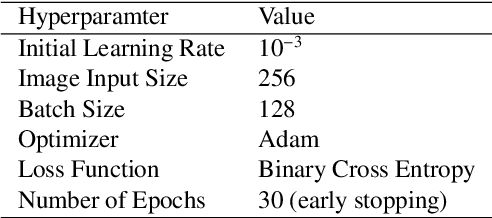
Abstract:Chest radiography is the most common radiographic examination performed in daily clinical practice for the detection of various heart and lung abnormalities. The large amount of data to be read and reported, with more than 100 studies per day for a single radiologist, poses a challenge in consistently maintaining high interpretation accuracy. The introduction of large-scale public datasets has led to a series of novel systems for automated abnormality classification. However, the labels of these datasets were obtained using natural language processed medical reports, yielding a large degree of label noise that can impact the performance. In this study, we propose novel training strategies that handle label noise from such suboptimal data. Prior label probabilities were measured on a subset of training data re-read by 4 board-certified radiologists and were used during training to increase the robustness of the training model to the label noise. Furthermore, we exploit the high comorbidity of abnormalities observed in chest radiography and incorporate this information to further reduce the impact of label noise. Additionally, anatomical knowledge is incorporated by training the system to predict lung and heart segmentation, as well as spatial knowledge labels. To deal with multiple datasets and images derived from various scanners that apply different post-processing techniques, we introduce a novel image normalization strategy. Experiments were performed on an extensive collection of 297,541 chest radiographs from 86,876 patients, leading to a state-of-the-art performance level for 17 abnormalities from 2 datasets. With an average AUC score of 0.880 across all abnormalities, our proposed training strategies can be used to significantly improve performance scores.
Automated detection and quantification of COVID-19 airspace disease on chest radiographs: A novel approach achieving radiologist-level performance using a CNN trained on digital reconstructed radiographs (DRRs) from CT-based ground-truth
Aug 13, 2020



Abstract:Purpose: To leverage volumetric quantification of airspace disease (AD) derived from a superior modality (CT) serving as ground truth, projected onto digitally reconstructed radiographs (DRRs) to: 1) train a convolutional neural network to quantify airspace disease on paired CXRs; and 2) compare the DRR-trained CNN to expert human readers in the CXR evaluation of patients with confirmed COVID-19. Materials and Methods: We retrospectively selected a cohort of 86 COVID-19 patients (with positive RT-PCR), from March-May 2020 at a tertiary hospital in the northeastern USA, who underwent chest CT and CXR within 48 hrs. The ground truth volumetric percentage of COVID-19 related AD (POv) was established by manual AD segmentation on CT. The resulting 3D masks were projected into 2D anterior-posterior digitally reconstructed radiographs (DRR) to compute area-based AD percentage (POa). A convolutional neural network (CNN) was trained with DRR images generated from a larger-scale CT dataset of COVID-19 and non-COVID-19 patients, automatically segmenting lungs, AD and quantifying POa on CXR. CNN POa results were compared to POa quantified on CXR by two expert readers and to the POv ground-truth, by computing correlations and mean absolute errors. Results: Bootstrap mean absolute error (MAE) and correlations between POa and POv were 11.98% [11.05%-12.47%] and 0.77 [0.70-0.82] for average of expert readers, and 9.56%-9.78% [8.83%-10.22%] and 0.78-0.81 [0.73-0.85] for the CNN, respectively. Conclusion: Our CNN trained with DRR using CT-derived airspace quantification achieved expert radiologist level of accuracy in the quantification of airspace disease on CXR, in patients with positive RT-PCR for COVID-19.
Extracting and Leveraging Nodule Features with Lung Inpainting for Local Feature Augmentation
Aug 05, 2020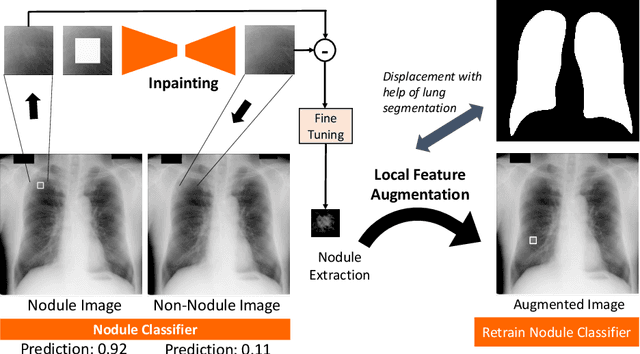
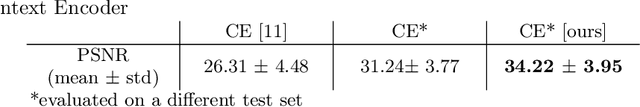
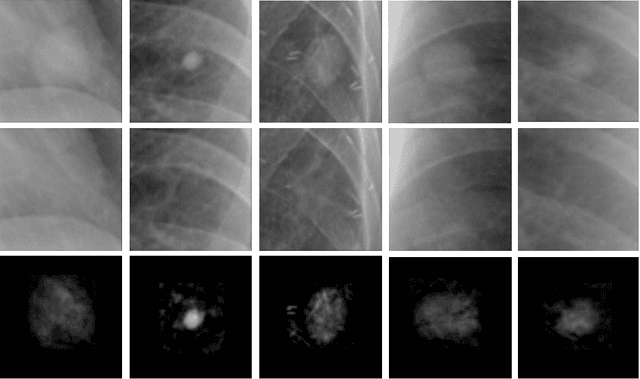
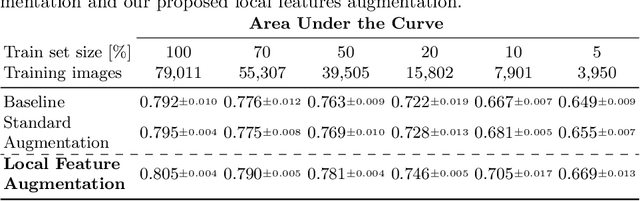
Abstract:Chest X-ray (CXR) is the most common examination for fast detection of pulmonary abnormalities. Recently, automated algorithms have been developed to classify multiple diseases and abnormalities in CXR scans. However, because of the limited availability of scans containing nodules and the subtle properties of nodules in CXRs, state-of-the-art methods do not perform well on nodule classification. To create additional data for the training process, standard augmentation techniques are applied. However, the variance introduced by these methods are limited as the images are typically modified globally. In this paper, we propose a method for local feature augmentation by extracting local nodule features using a generative inpainting network. The network is applied to generate realistic, healthy tissue and structures in patches containing nodules. The nodules are entirely removed in the inpainted representation. The extraction of the nodule features is processed by subtraction of the inpainted patch from the nodule patch. With arbitrary displacement of the extracted nodules in the lung area across different CXR scans and further local modifications during training, we significantly increase the nodule classification performance and outperform state-of-the-art augmentation methods.
Quantifying and Leveraging Predictive Uncertainty for Medical Image Assessment
Jul 08, 2020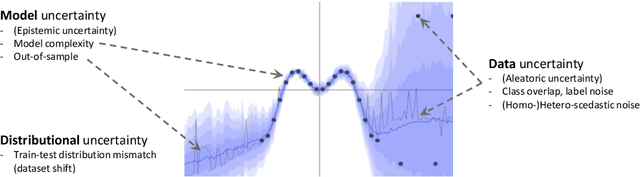

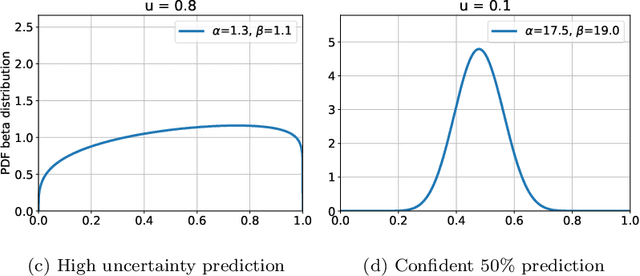
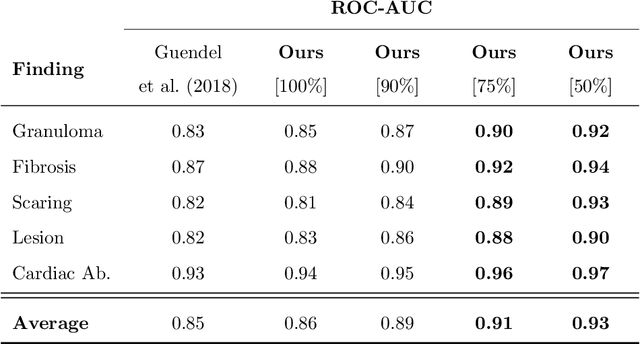
Abstract:The interpretation of medical images is a challenging task, often complicated by the presence of artifacts, occlusions, limited contrast and more. Most notable is the case of chest radiography, where there is a high inter-rater variability in the detection and classification of abnormalities. This is largely due to inconclusive evidence in the data or subjective definitions of disease appearance. An additional example is the classification of anatomical views based on 2D Ultrasound images. Often, the anatomical context captured in a frame is not sufficient to recognize the underlying anatomy. Current machine learning solutions for these problems are typically limited to providing probabilistic predictions, relying on the capacity of underlying models to adapt to limited information and the high degree of label noise. In practice, however, this leads to overconfident systems with poor generalization on unseen data. To account for this, we propose a system that learns not only the probabilistic estimate for classification, but also an explicit uncertainty measure which captures the confidence of the system in the predicted output. We argue that this approach is essential to account for the inherent ambiguity characteristic of medical images from different radiologic exams including computed radiography, ultrasonography and magnetic resonance imaging. In our experiments we demonstrate that sample rejection based on the predicted uncertainty can significantly improve the ROC-AUC for various tasks, e.g., by 8% to 0.91 with an expected rejection rate of under 25% for the classification of different abnormalities in chest radiographs. In addition, we show that using uncertainty-driven bootstrapping to filter the training data, one can achieve a significant increase in robustness and accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge