Xiaowei Xu
Myocardial Segmentation of Cardiac MRI Sequences with Temporal Consistency for Coronary Artery Disease Diagnosis
Dec 29, 2020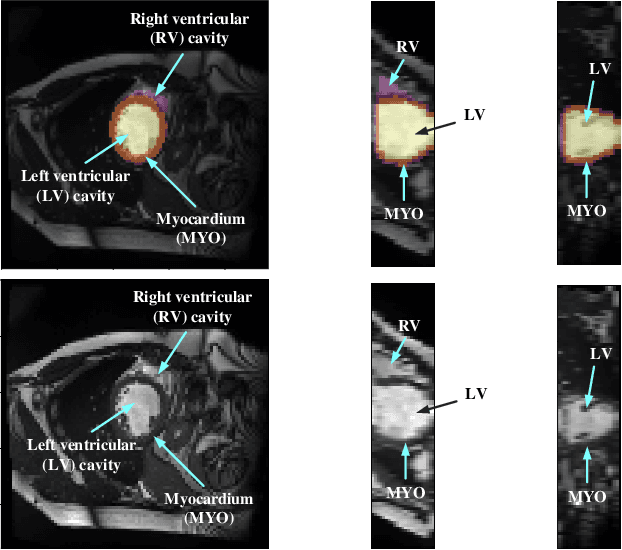
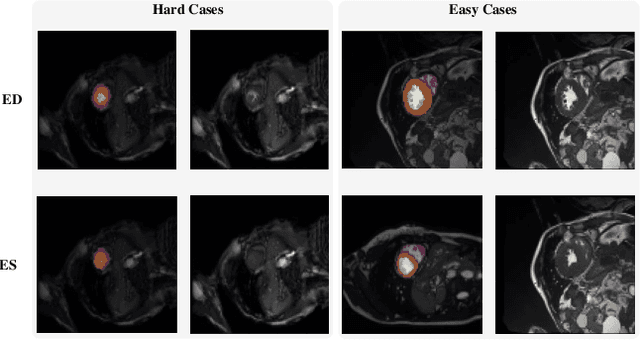
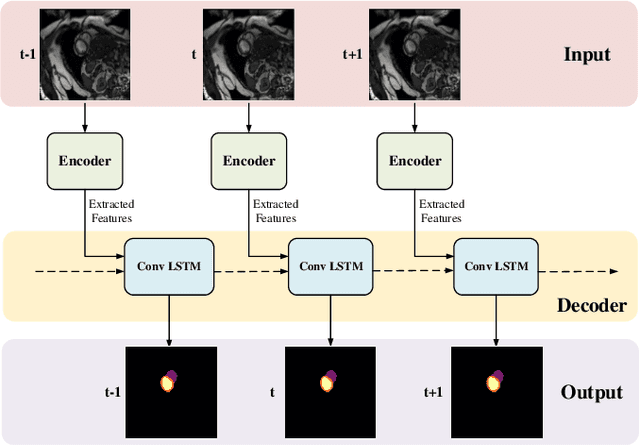
Abstract:Coronary artery disease (CAD) is the most common cause of death globally, and its diagnosis is usually based on manual myocardial segmentation of Magnetic Resonance Imaging (MRI) sequences. As the manual segmentation is tedious, time-consuming and with low applicability, automatic myocardial segmentation using machine learning techniques has been widely explored recently. However, almost all the existing methods treat the input MRI sequences independently, which fails to capture the temporal information between sequences, e.g., the shape and location information of the myocardium in sequences along time. In this paper, we propose a myocardial segmentation framework for sequence of cardiac MRI (CMR) scanning images of left ventricular cavity, right ventricular cavity, and myocardium. Specifically, we propose to combine conventional networks and recurrent networks to incorporate temporal information between sequences to ensure temporal consistent. We evaluated our framework on the Automated Cardiac Diagnosis Challenge (ACDC) dataset. Experiment results demonstrate that our framework can improve the segmentation accuracy by up to 2% in Dice coefficient.
C2F-FWN: Coarse-to-Fine Flow Warping Network for Spatial-Temporal Consistent Motion Transfer
Dec 16, 2020
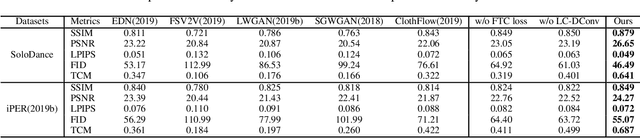
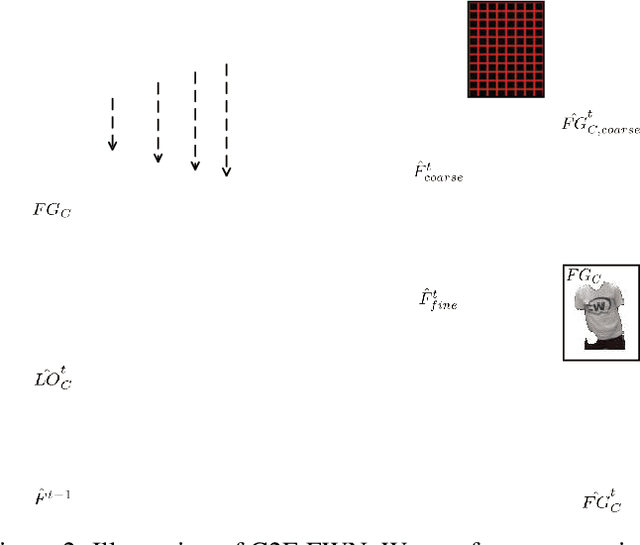
Abstract:Human video motion transfer (HVMT) aims to synthesize videos that one person imitates other persons' actions. Although existing GAN-based HVMT methods have achieved great success, they either fail to preserve appearance details due to the loss of spatial consistency between synthesized and exemplary images, or generate incoherent video results due to the lack of temporal consistency among video frames. In this paper, we propose Coarse-to-Fine Flow Warping Network (C2F-FWN) for spatial-temporal consistent HVMT. Particularly, C2F-FWN utilizes coarse-to-fine flow warping and Layout-Constrained Deformable Convolution (LC-DConv) to improve spatial consistency, and employs Flow Temporal Consistency (FTC) Loss to enhance temporal consistency. In addition, provided with multi-source appearance inputs, C2F-FWN can support appearance attribute editing with great flexibility and efficiency. Besides public datasets, we also collected a large-scale HVMT dataset named SoloDance for evaluation. Extensive experiments conducted on our SoloDance dataset and the iPER dataset show that our approach outperforms state-of-art HVMT methods in terms of both spatial and temporal consistency. Source code and the SoloDance dataset are available at https://github.com/wswdx/C2F-FWN.
Weakly-Supervised Cross-Domain Adaptation for Endoscopic Lesions Segmentation
Dec 08, 2020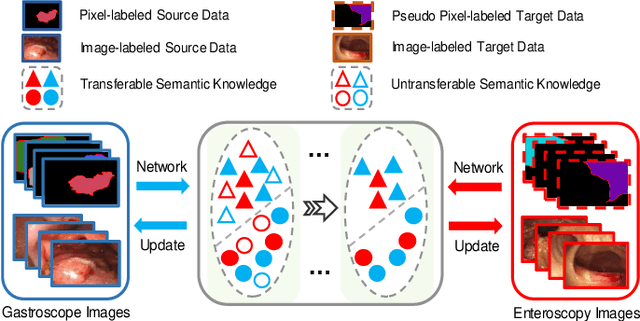
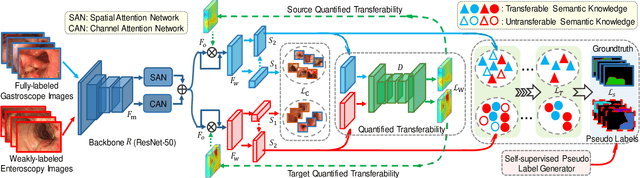
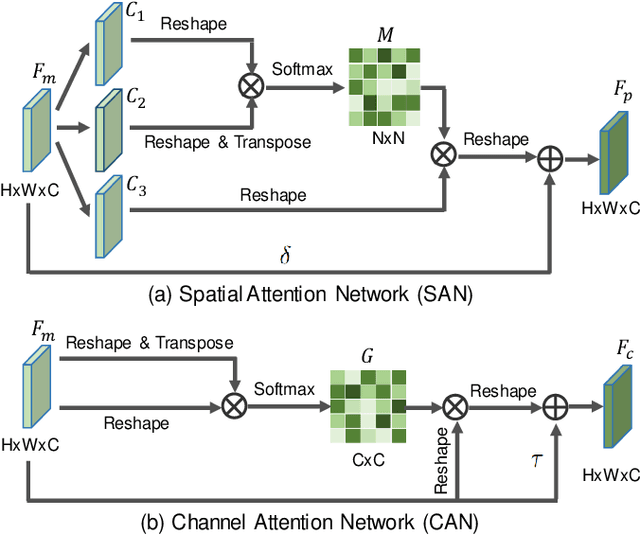
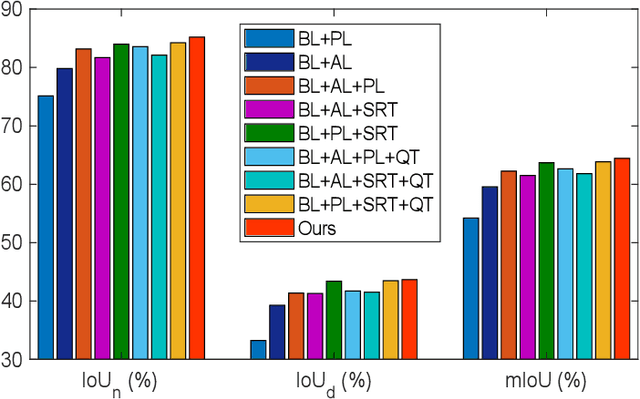
Abstract:Weakly-supervised learning has attracted growing research attention on medical lesions segmentation due to significant saving in pixel-level annotation cost. However, 1) most existing methods require effective prior and constraints to explore the intrinsic lesions characterization, which only generates incorrect and rough prediction; 2) they neglect the underlying semantic dependencies among weakly-labeled target enteroscopy diseases and fully-annotated source gastroscope lesions, while forcefully utilizing untransferable dependencies leads to the negative performance. To tackle above issues, we propose a new weakly-supervised lesions transfer framework, which can not only explore transferable domain-invariant knowledge across different datasets, but also prevent the negative transfer of untransferable representations. Specifically, a Wasserstein quantified transferability framework is developed to highlight widerange transferable contextual dependencies, while neglecting the irrelevant semantic characterizations. Moreover, a novel selfsupervised pseudo label generator is designed to equally provide confident pseudo pixel labels for both hard-to-transfer and easyto-transfer target samples. It inhibits the enormous deviation of false pseudo pixel labels under the self-supervision manner. Afterwards, dynamically-searched feature centroids are aligned to narrow category-wise distribution shift. Comprehensive theoretical analysis and experiments show the superiority of our model on the endoscopic dataset and several public datasets.
Do Noises Bother Human and Neural Networks In the Same Way? A Medical Image Analysis Perspective
Nov 04, 2020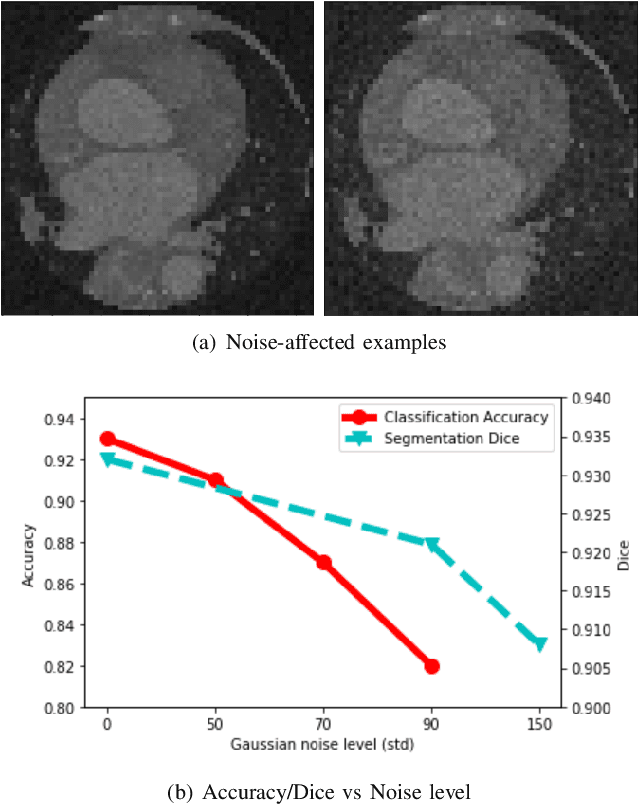
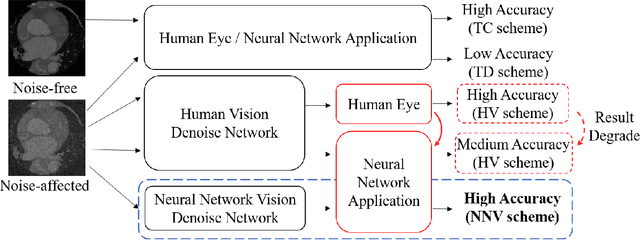
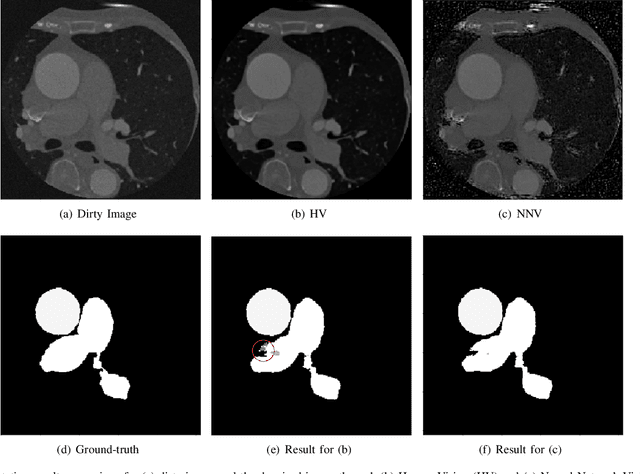
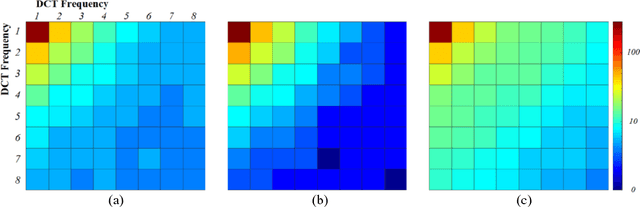
Abstract:Deep learning had already demonstrated its power in medical images, including denoising, classification, segmentation, etc. All these applications are proposed to automatically analyze medical images beforehand, which brings more information to radiologists during clinical assessment for accuracy improvement. Recently, many medical denoising methods had shown their significant artifact reduction result and noise removal both quantitatively and qualitatively. However, those existing methods are developed around human-vision, i.e., they are designed to minimize the noise effect that can be perceived by human eyes. In this paper, we introduce an application-guided denoising framework, which focuses on denoising for the following neural networks. In our experiments, we apply the proposed framework to different datasets, models, and use cases. Experimental results show that our proposed framework can achieve a better result than human-vision denoising network.
CSCL: Critical Semantic-Consistent Learning for Unsupervised Domain Adaptation
Aug 24, 2020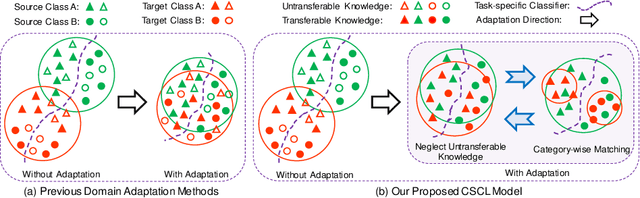
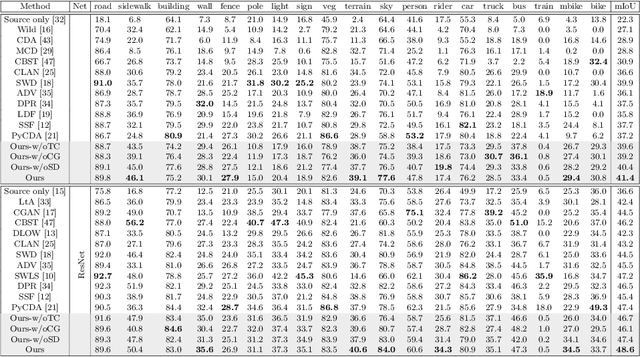
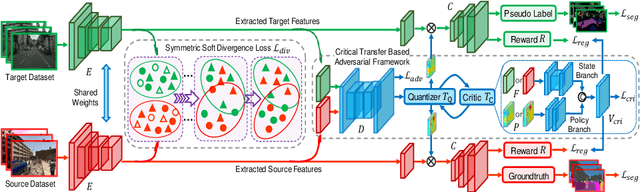
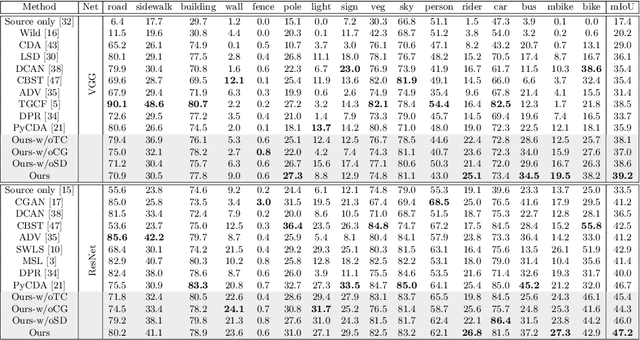
Abstract:Unsupervised domain adaptation without consuming annotation process for unlabeled target data attracts appealing interests in semantic segmentation. However, 1) existing methods neglect that not all semantic representations across domains are transferable, which cripples domain-wise transfer with untransferable knowledge; 2) they fail to narrow category-wise distribution shift due to category-agnostic feature alignment. To address above challenges, we develop a new Critical Semantic-Consistent Learning (CSCL) model, which mitigates the discrepancy of both domain-wise and category-wise distributions. Specifically, a critical transfer based adversarial framework is designed to highlight transferable domain-wise knowledge while neglecting untransferable knowledge. Transferability-critic guides transferability-quantizer to maximize positive transfer gain under reinforcement learning manner, although negative transfer of untransferable knowledge occurs. Meanwhile, with the help of confidence-guided pseudo labels generator of target samples, a symmetric soft divergence loss is presented to explore inter-class relationships and facilitate category-wise distribution alignment. Experiments on several datasets demonstrate the superiority of our model.
Towards Cardiac Intervention Assistance: Hardware-aware Neural Architecture Exploration for Real-Time 3D Cardiac Cine MRI Segmentation
Aug 17, 2020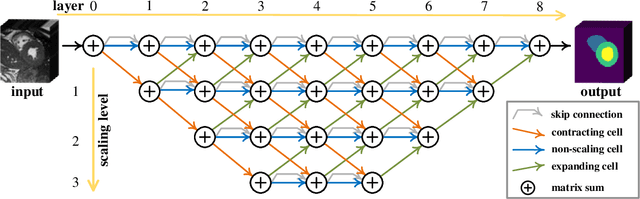
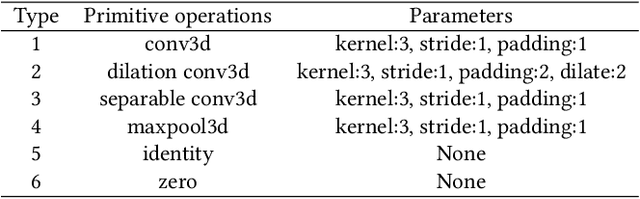
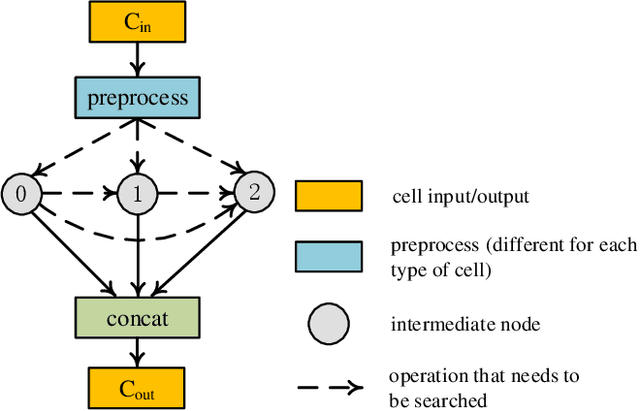
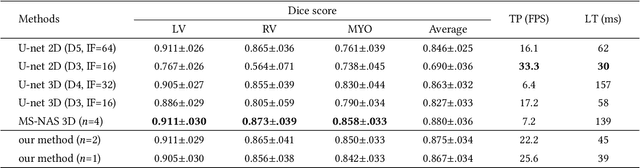
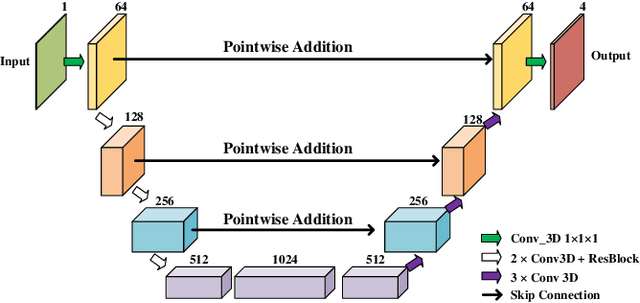
Abstract:Real-time cardiac magnetic resonance imaging (MRI) plays an increasingly important role in guiding various cardiac interventions. In order to provide better visual assistance, the cine MRI frames need to be segmented on-the-fly to avoid noticeable visual lag. In addition, considering reliability and patient data privacy, the computation is preferably done on local hardware. State-of-the-art MRI segmentation methods mostly focus on accuracy only, and can hardly be adopted for real-time application or on local hardware. In this work, we present the first hardware-aware multi-scale neural architecture search (NAS) framework for real-time 3D cardiac cine MRI segmentation. The proposed framework incorporates a latency regularization term into the loss function to handle real-time constraints, with the consideration of underlying hardware. In addition, the formulation is fully differentiable with respect to the architecture parameters, so that stochastic gradient descent (SGD) can be used for optimization to reduce the computation cost while maintaining optimization quality. Experimental results on ACDC MICCAI 2017 dataset demonstrate that our hardware-aware multi-scale NAS framework can reduce the latency by up to 3.5 times and satisfy the real-time constraints, while still achieving competitive segmentation accuracy, compared with the state-of-the-art NAS segmentation framework.
ICA-UNet: ICA Inspired Statistical UNet for Real-time 3D Cardiac Cine MRI Segmentation
Jul 18, 2020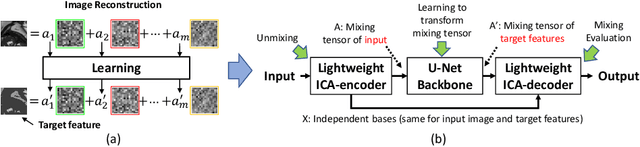
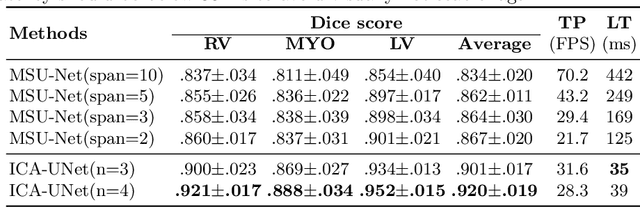
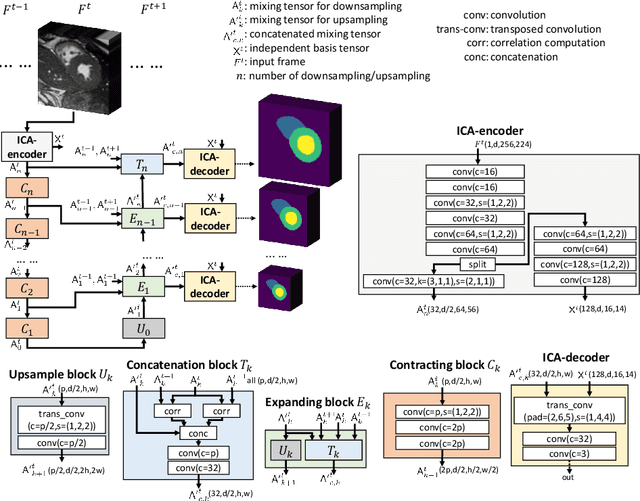
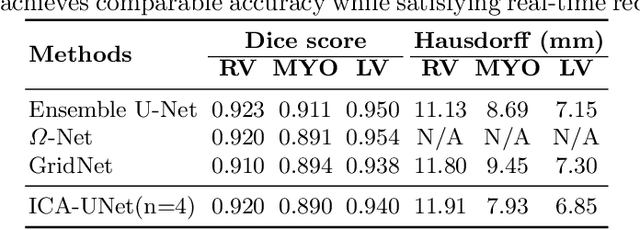
Abstract:Real-time cine magnetic resonance imaging (MRI) plays an increasingly important role in various cardiac interventions. In order to enable fast and accurate visual assistance, the temporal frames need to be segmented on-the-fly. However, state-of-the-art MRI segmentation methods are used either offline because of their high computation complexity, or in real-time but with significant accuracy loss and latency increase (causing visually noticeable lag). As such, they can hardly be adopted to assist visual guidance. In this work, inspired by a new interpretation of Independent Component Analysis (ICA) for learning, we propose a novel ICA-UNet for real-time 3D cardiac cine MRI segmentation. Experiments using the MICCAI ACDC 2017 dataset show that, compared with the state-of-the-arts, ICA-UNet not only achieves higher Dice scores, but also meets the real-time requirements for both throughput and latency (up to 12.6X reduction), enabling real-time guidance for cardiac interventions without visual lag.
BUNET: Blind Medical Image Segmentation Based on Secure UNET
Jul 14, 2020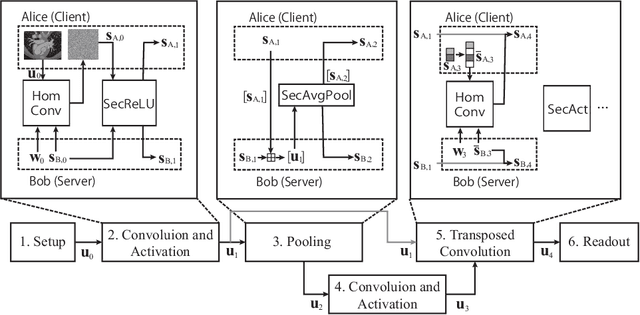
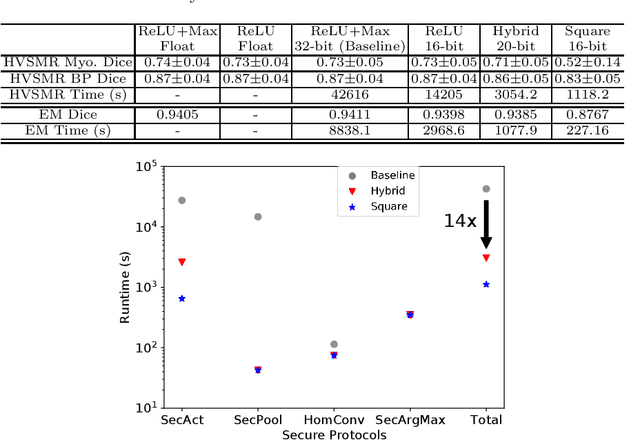
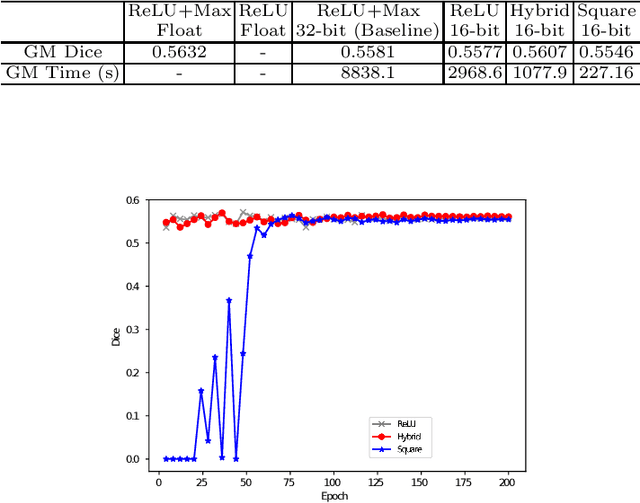
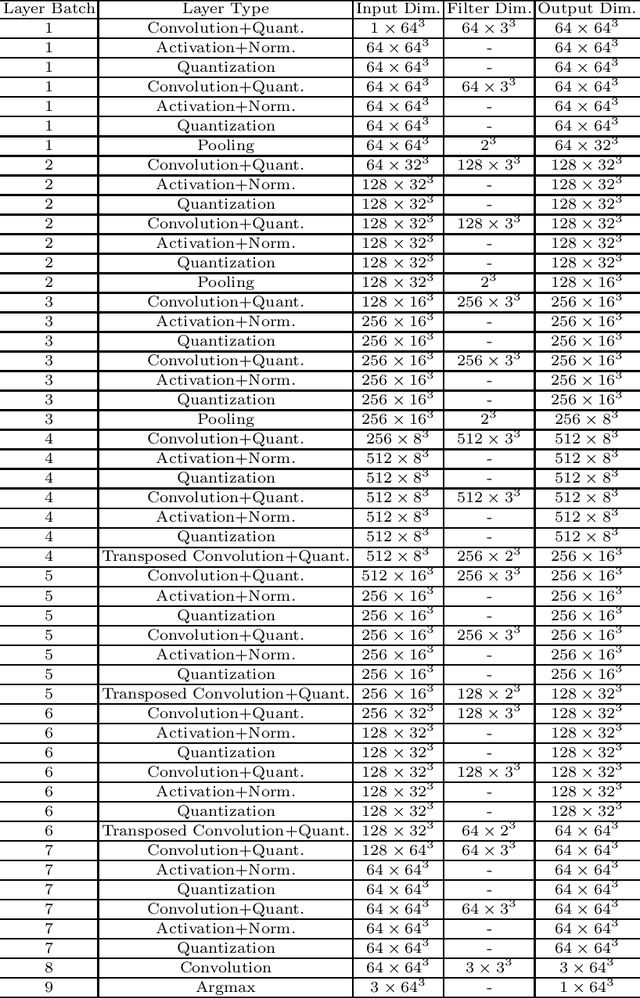
Abstract:The strict security requirements placed on medical records by various privacy regulations become major obstacles in the age of big data. To ensure efficient machine learning as a service schemes while protecting data confidentiality, in this work, we propose blind UNET (BUNET), a secure protocol that implements privacy-preserving medical image segmentation based on the UNET architecture. In BUNET, we efficiently utilize cryptographic primitives such as homomorphic encryption and garbled circuits (GC) to design a complete secure protocol for the UNET neural architecture. In addition, we perform extensive architectural search in reducing the computational bottleneck of GC-based secure activation protocols with high-dimensional input data. In the experiment, we thoroughly examine the parameter space of our protocol, and show that we can achieve up to 14x inference time reduction compared to the-state-of-the-art secure inference technique on a baseline architecture with negligible accuracy degradation.
Evolving Metric Learning for Incremental and Decremental Features
Jun 27, 2020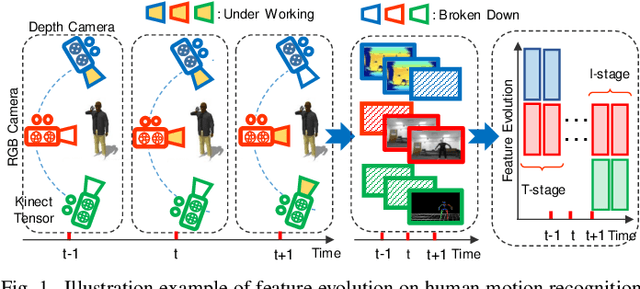
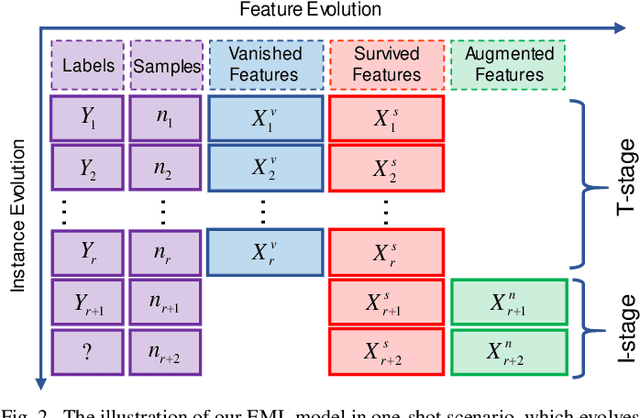
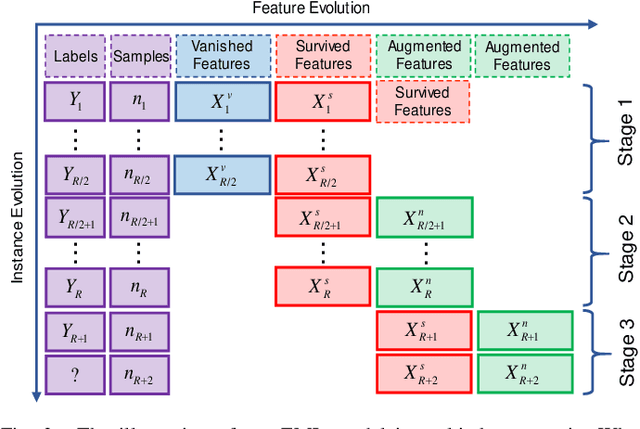

Abstract:Online metric learning has been widely exploited for large-scale data classification due to the low computational cost. However, amongst online practical scenarios where the features are evolving (e.g., some features are vanished and some new features are augmented), most metric learning models cannot be successfully applied into these scenarios although they can tackle the evolving instances efficiently. To address the challenge, we propose a new online Evolving Metric Learning (EML) model for incremental and decremental features, which can handle the instance and feature evolutions simultaneously by incorporating with a smoothed Wasserstein metric distance. Specifically, our model contains two essential stages: the Transforming stage (T-stage) and the Inheriting stage (I-stage). For the T-stage, we propose to extract important information from vanished features while neglecting non-informative knowledge, and forward it into survived features by transforming them into a low-rank discriminative metric space. It further explores the intrinsic low-rank structure of heterogeneous samples to reduce the computation and memory burden especially for highly-dimensional large-scale data. For the I-stage, we inherit the metric performance of survived features from the T-stage and then expand to include the augmented new features. Moreover, the smoothed Wasserstein distance is utilized to characterize the similarity relations among the complex and heterogeneous data, since the evolving features in the different stages are not strictly aligned. In addition to tackling the challenges in one-shot case, we also extend our model into multi-shot scenario. After deriving an efficient optimization method for both T-stage and I-stage, extensive experiments on several benchmark datasets verify the superiority of our model.
What Can Be Transferred: Unsupervised Domain Adaptation for Endoscopic Lesions Segmentation
Apr 24, 2020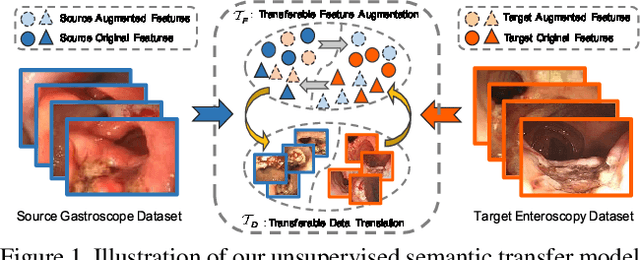
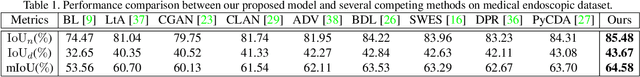
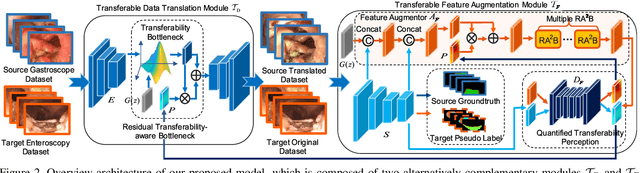
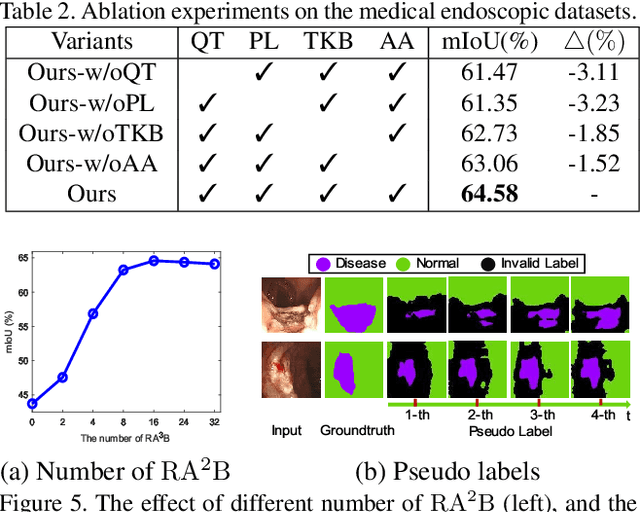
Abstract:Unsupervised domain adaptation has attracted growing research attention on semantic segmentation. However, 1) most existing models cannot be directly applied into lesions transfer of medical images, due to the diverse appearances of same lesion among different datasets; 2) equal attention has been paid into all semantic representations instead of neglecting irrelevant knowledge, which leads to negative transfer of untransferable knowledge. To address these challenges, we develop a new unsupervised semantic transfer model including two complementary modules (i.e., T_D and T_F ) for endoscopic lesions segmentation, which can alternatively determine where and how to explore transferable domain-invariant knowledge between labeled source lesions dataset (e.g., gastroscope) and unlabeled target diseases dataset (e.g., enteroscopy). Specifically, T_D focuses on where to translate transferable visual information of medical lesions via residual transferability-aware bottleneck, while neglecting untransferable visual characterizations. Furthermore, T_F highlights how to augment transferable semantic features of various lesions and automatically ignore untransferable representations, which explores domain-invariant knowledge and in return improves the performance of T_D. To the end, theoretical analysis and extensive experiments on medical endoscopic dataset and several non-medical public datasets well demonstrate the superiority of our proposed model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge