Tianchen Wang
SPAR: Self-supervised Placement-Aware Representation Learning for Multi-Node IoT Systems
May 22, 2025Abstract:This work develops the underpinnings of self-supervised placement-aware representation learning given spatially-distributed (multi-view and multimodal) sensor observations, motivated by the need to represent external environmental state in multi-sensor IoT systems in a manner that correctly distills spatial phenomena from the distributed multi-vantage observations. The objective of sensing in IoT systems is, in general, to collectively represent an externally observed environment given multiple vantage points from which sensory observations occur. Pretraining of models that help interpret sensor data must therefore encode the relation between signals observed by sensors and the observers' vantage points in order to attain a representation that encodes the observed spatial phenomena in a manner informed by the specific placement of the measuring instruments, while allowing arbitrary placement. The work significantly advances self-supervised model pretraining from IoT signals beyond current solutions that often overlook the distinctive spatial nature of IoT data. Our framework explicitly learns the dependencies between measurements and geometric observer layouts and structural characteristics, guided by a core design principle: the duality between signals and observer positions. We further provide theoretical analyses from the perspectives of information theory and occlusion-invariant representation learning to offer insight into the rationale behind our design. Experiments on three real-world datasets--covering vehicle monitoring, human activity recognition, and earthquake localization--demonstrate the superior generalizability and robustness of our method across diverse modalities, sensor placements, application-level inference tasks, and spatial scales.
Lightweight Delivery Detection on Doorbell Cameras
May 13, 2023Abstract:Despite recent advances in video-based action recognition and robust spatio-temporal modeling, most of the proposed approaches rely on the abundance of computational resources to afford running huge and computation-intensive convolutional or transformer-based neural networks to obtain satisfactory results. This limits the deployment of such models on edge devices with limited power and computing resources. In this work we investigate an important smart home application, video based delivery detection, and present a simple and lightweight pipeline for this task that can run on resource-constrained doorbell cameras. Our proposed pipeline relies on motion cues to generate a set of coarse activity proposals followed by their classification with a mobile-friendly 3DCNN network. For training we design a novel semi-supervised attention module that helps the network to learn robust spatio-temporal features and adopt an evidence-based optimization objective that allows for quantifying the uncertainty of predictions made by the network. Experimental results on our curated delivery dataset shows the significant effectiveness of our pipeline compared to alternatives and highlights the benefits of our training phase novelties to achieve free and considerable inference-time performance gains.
ImageCAS: A Large-Scale Dataset and Benchmark for Coronary Artery Segmentation based on Computed Tomography Angiography Images
Nov 03, 2022Abstract:Cardiovascular disease (CVD) accounts for about half of non-communicable diseases. Vessel stenosis in the coronary artery is considered to be the major risk of CVD. Computed tomography angiography (CTA) is one of the widely used noninvasive imaging modalities in coronary artery diagnosis due to its superior image resolution. Clinically, segmentation of coronary arteries is essential for the diagnosis and quantification of coronary artery disease. Recently, a variety of works have been proposed to address this problem. However, on one hand, most works rely on in-house datasets, and only a few works published their datasets to the public which only contain tens of images. On the other hand, their source code have not been published, and most follow-up works have not made comparison with existing works, which makes it difficult to judge the effectiveness of the methods and hinders the further exploration of this challenging yet critical problem in the community. In this paper, we propose a large-scale dataset for coronary artery segmentation on CTA images. In addition, we have implemented a benchmark in which we have tried our best to implement several typical existing methods. Furthermore, we propose a strong baseline method which combines multi-scale patch fusion and two-stage processing to extract the details of vessels. Comprehensive experiments show that the proposed method achieves better performance than existing works on the proposed large-scale dataset. The benchmark and the dataset are published at https://github.com/XiaoweiXu/ImageCAS-A-Large-Scale-Dataset-and-Benchmark-for-Coronary-Artery-Segmentation-based-on-CT.
ImageTBAD: A 3D Computed Tomography Angiography Image Dataset for Automatic Segmentation of Type-B Aortic Dissection
Sep 01, 2021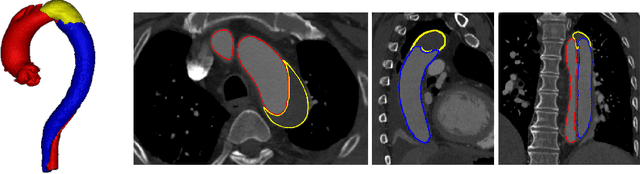

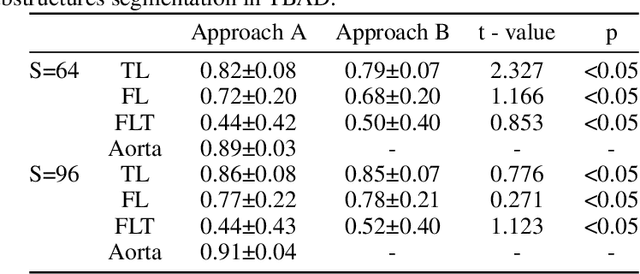
Abstract:Type-B Aortic Dissection (TBAD) is one of the most serious cardiovascular events characterized by a growing yearly incidence,and the severity of disease prognosis. Currently, computed tomography angiography (CTA) has been widely adopted for the diagnosis and prognosis of TBAD. Accurate segmentation of true lumen (TL), false lumen (FL), and false lumen thrombus (FLT) in CTA are crucial for the precise quantification of anatomical features. However, existing works only focus on only TL and FL without considering FLT. In this paper, we propose ImageTBAD, the first 3D computed tomography angiography (CTA) image dataset of TBAD with annotation of TL, FL, and FLT. The proposed dataset contains 100 TBAD CTA images, which is of decent size compared with existing medical imaging datasets. As FLT can appear almost anywhere along the aorta with irregular shapes, segmentation of FLT presents a wide class of segmentation problems where targets exist in a variety of positions with irregular shapes. We further propose a baseline method for automatic segmentation of TBAD. Results show that the baseline method can achieve comparable results with existing works on aorta and TL segmentation. However, the segmentation accuracy of FLT is only 52%, which leaves large room for improvement and also shows the challenge of our dataset. To facilitate further research on this challenging problem, our dataset and codes are released to the public.
EchoCP: An Echocardiography Dataset in Contrast Transthoracic Echocardiography for Patent Foramen Ovale Diagnosis
May 18, 2021
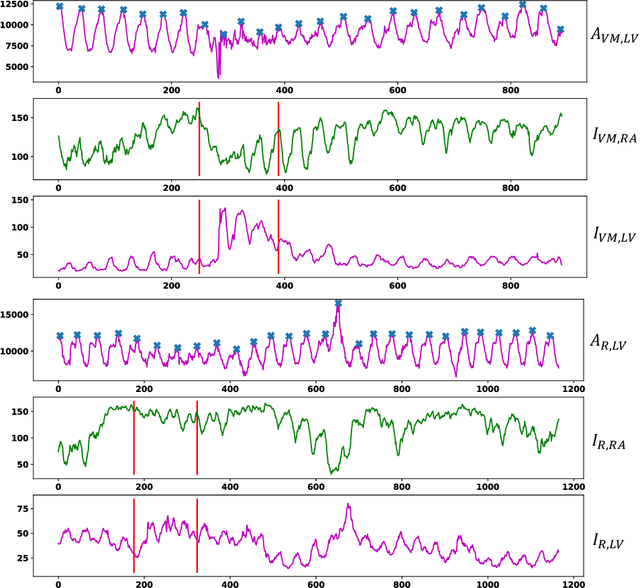
Abstract:Patent foramen ovale (PFO) is a potential separation between the septum, primum and septum secundum located in the anterosuperior portion of the atrial septum. PFO is one of the main factors causing cryptogenic stroke which is the fifth leading cause of death in the United States. For PFO diagnosis, contrast transthoracic echocardiography (cTTE) is preferred as being a more robust method compared with others. However, the current PFO diagnosis through cTTE is extremely slow as it is proceeded manually by sonographers on echocardiography videos. Currently there is no publicly available dataset for this important topic in the community. In this paper, we present EchoCP, as the first echocardiography dataset in cTTE targeting PFO diagnosis. EchoCP consists of 30 patients with both rest and Valsalva maneuver videos which covers various PFO grades. We further establish an automated baseline method for PFO diagnosis based on the state-of-the-art cardiac chamber segmentation technique, which achieves 0.89 average mean Dice score, but only 0.70/0.67 mean accuracies for PFO diagnosis, leaving large room for improvement. We hope that the challenging EchoCP dataset can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released.
Quantization of Deep Neural Networks for Accurate EdgeComputing
Apr 25, 2021
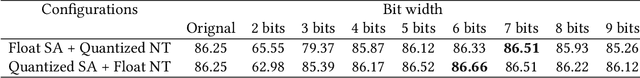
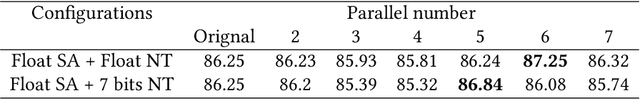
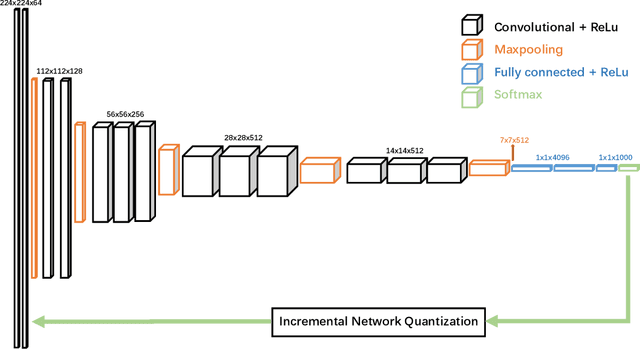
Abstract:Deep neural networks (DNNs) have demonstrated their great potential in recent years, exceeding the per-formance of human experts in a wide range of applications. Due to their large sizes, however, compressiontechniques such as weight quantization and pruning are usually applied before they can be accommodated onthe edge. It is generally believed that quantization leads to performance degradation, and plenty of existingworks have explored quantization strategies aiming at minimum accuracy loss. In this paper, we argue thatquantization, which essentially imposes regularization on weight representations, can sometimes help toimprove accuracy. We conduct comprehensive experiments on three widely used applications: fully con-nected network (FCN) for biomedical image segmentation, convolutional neural network (CNN) for imageclassification on ImageNet, and recurrent neural network (RNN) for automatic speech recognition, and experi-mental results show that quantization can improve the accuracy by 1%, 1.95%, 4.23% on the three applicationsrespectively with 3.5x-6.4x memory reduction.
Multi-Cycle-Consistent Adversarial Networks for Edge Denoising of Computed Tomography Images
Apr 25, 2021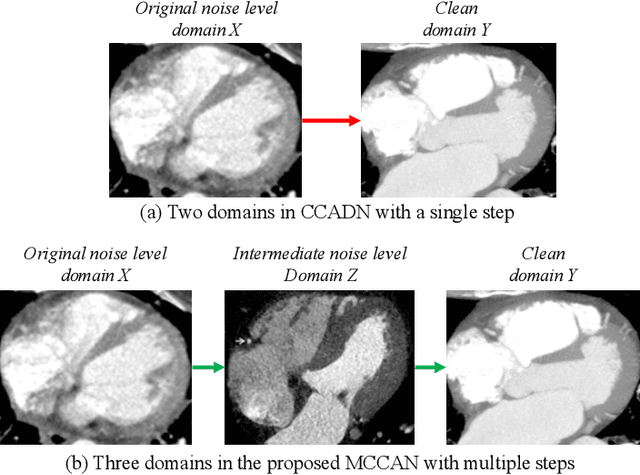
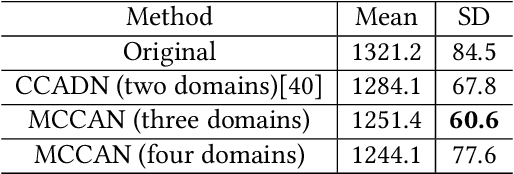
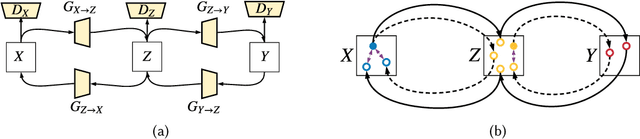
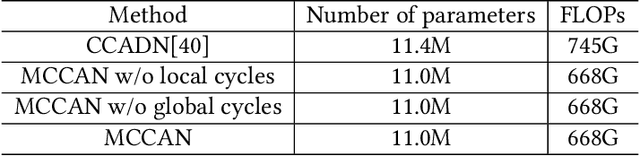
Abstract:As one of the most commonly ordered imaging tests, computed tomography (CT) scan comes with inevitable radiation exposure that increases the cancer risk to patients. However, CT image quality is directly related to radiation dose, thus it is desirable to obtain high-quality CT images with as little dose as possible. CT image denoising tries to obtain high dose like high-quality CT images (domain X) from low dose low-quality CTimages (domain Y), which can be treated as an image-to-image translation task where the goal is to learn the transform between a source domain X (noisy images) and a target domain Y (clean images). In this paper, we propose a multi-cycle-consistent adversarial network (MCCAN) that builds intermediate domains and enforces both local and global cycle-consistency for edge denoising of CT images. The global cycle-consistency couples all generators together to model the whole denoising process, while the local cycle-consistency imposes effective supervision on the process between adjacent domains. Experiments show that both local and global cycle-consistency are important for the success of MCCAN, which outperformsCCADN in terms of denoising quality with slightly less computation resource consumption.
ImageCHD: A 3D Computed Tomography Image Dataset for Classification of Congenital Heart Disease
Jan 26, 2021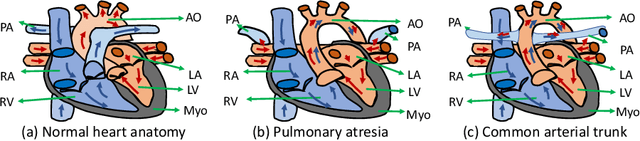
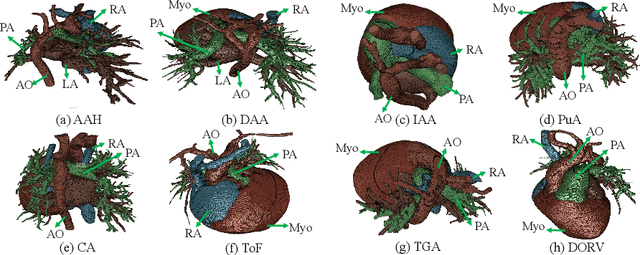
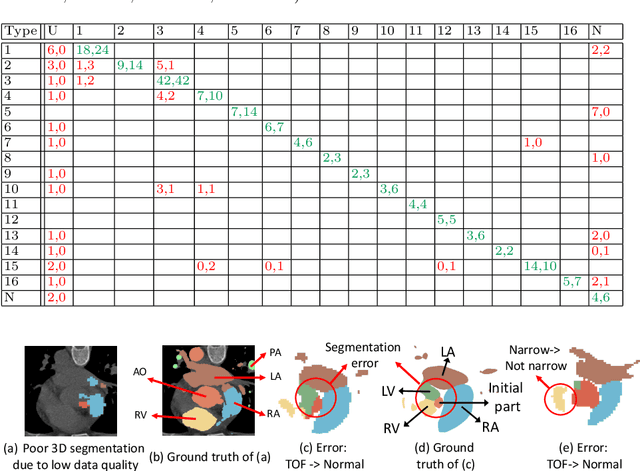
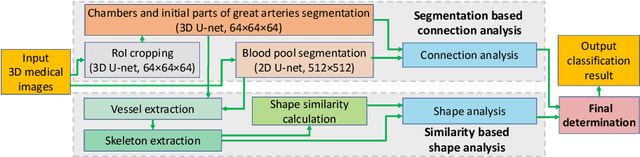
Abstract:Congenital heart disease (CHD) is the most common type of birth defect, which occurs 1 in every 110 births in the United States. CHD usually comes with severe variations in heart structure and great artery connections that can be classified into many types. Thus highly specialized domain knowledge and the time-consuming human process is needed to analyze the associated medical images. On the other hand, due to the complexity of CHD and the lack of dataset, little has been explored on the automatic diagnosis (classification) of CHDs. In this paper, we present ImageCHD, the first medical image dataset for CHD classification. ImageCHD contains 110 3D Computed Tomography (CT) images covering most types of CHD, which is of decent size Classification of CHDs requires the identification of large structural changes without any local tissue changes, with limited data. It is an example of a larger class of problems that are quite difficult for current machine-learning-based vision methods to solve. To demonstrate this, we further present a baseline framework for the automatic classification of CHD, based on a state-of-the-art CHD segmentation method. Experimental results show that the baseline framework can only achieve a classification accuracy of 82.0\% under a selective prediction scheme with 88.4\% coverage, leaving big room for further improvement. We hope that ImageCHD can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released to the public compared with existing medical imaging datasets.
Towards Cardiac Intervention Assistance: Hardware-aware Neural Architecture Exploration for Real-Time 3D Cardiac Cine MRI Segmentation
Aug 17, 2020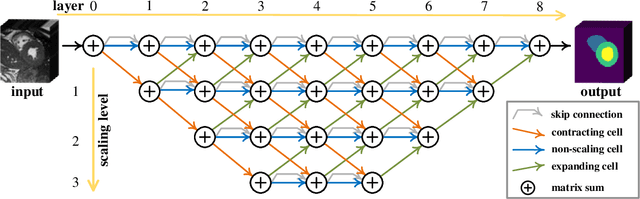
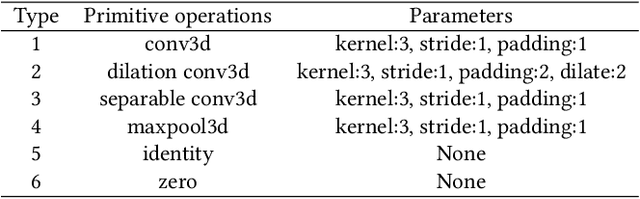
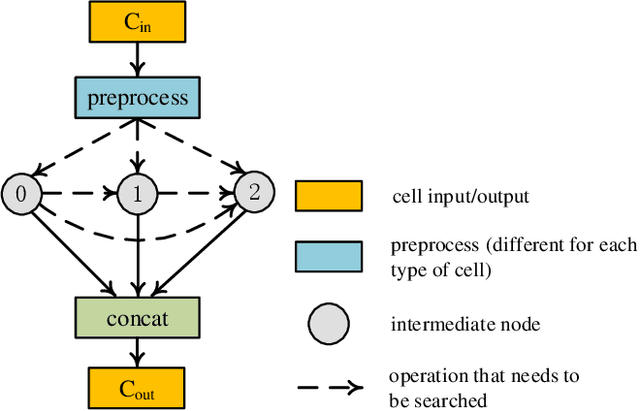
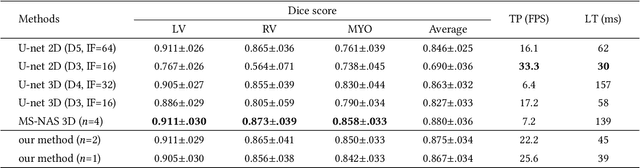
Abstract:Real-time cardiac magnetic resonance imaging (MRI) plays an increasingly important role in guiding various cardiac interventions. In order to provide better visual assistance, the cine MRI frames need to be segmented on-the-fly to avoid noticeable visual lag. In addition, considering reliability and patient data privacy, the computation is preferably done on local hardware. State-of-the-art MRI segmentation methods mostly focus on accuracy only, and can hardly be adopted for real-time application or on local hardware. In this work, we present the first hardware-aware multi-scale neural architecture search (NAS) framework for real-time 3D cardiac cine MRI segmentation. The proposed framework incorporates a latency regularization term into the loss function to handle real-time constraints, with the consideration of underlying hardware. In addition, the formulation is fully differentiable with respect to the architecture parameters, so that stochastic gradient descent (SGD) can be used for optimization to reduce the computation cost while maintaining optimization quality. Experimental results on ACDC MICCAI 2017 dataset demonstrate that our hardware-aware multi-scale NAS framework can reduce the latency by up to 3.5 times and satisfy the real-time constraints, while still achieving competitive segmentation accuracy, compared with the state-of-the-art NAS segmentation framework.
ICA-UNet: ICA Inspired Statistical UNet for Real-time 3D Cardiac Cine MRI Segmentation
Jul 18, 2020
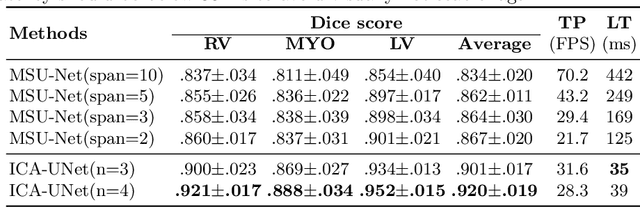
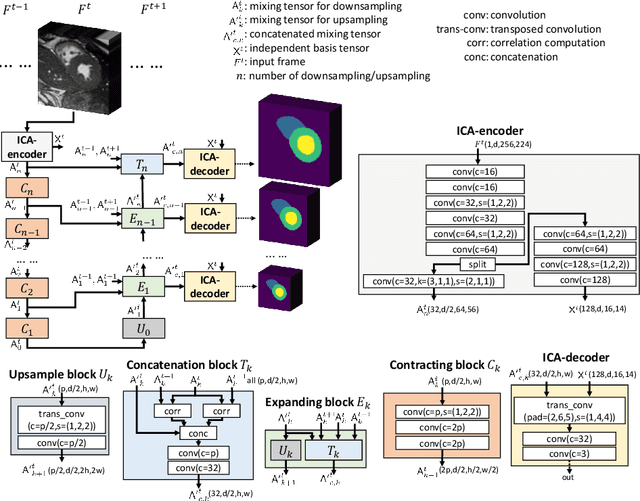
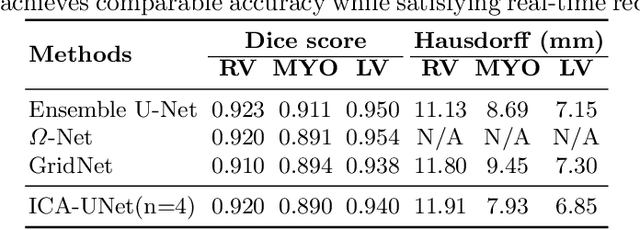
Abstract:Real-time cine magnetic resonance imaging (MRI) plays an increasingly important role in various cardiac interventions. In order to enable fast and accurate visual assistance, the temporal frames need to be segmented on-the-fly. However, state-of-the-art MRI segmentation methods are used either offline because of their high computation complexity, or in real-time but with significant accuracy loss and latency increase (causing visually noticeable lag). As such, they can hardly be adopted to assist visual guidance. In this work, inspired by a new interpretation of Independent Component Analysis (ICA) for learning, we propose a novel ICA-UNet for real-time 3D cardiac cine MRI segmentation. Experiments using the MICCAI ACDC 2017 dataset show that, compared with the state-of-the-arts, ICA-UNet not only achieves higher Dice scores, but also meets the real-time requirements for both throughput and latency (up to 12.6X reduction), enabling real-time guidance for cardiac interventions without visual lag.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge