Meiping Huang
ImageCAS: A Large-Scale Dataset and Benchmark for Coronary Artery Segmentation based on Computed Tomography Angiography Images
Nov 03, 2022Abstract:Cardiovascular disease (CVD) accounts for about half of non-communicable diseases. Vessel stenosis in the coronary artery is considered to be the major risk of CVD. Computed tomography angiography (CTA) is one of the widely used noninvasive imaging modalities in coronary artery diagnosis due to its superior image resolution. Clinically, segmentation of coronary arteries is essential for the diagnosis and quantification of coronary artery disease. Recently, a variety of works have been proposed to address this problem. However, on one hand, most works rely on in-house datasets, and only a few works published their datasets to the public which only contain tens of images. On the other hand, their source code have not been published, and most follow-up works have not made comparison with existing works, which makes it difficult to judge the effectiveness of the methods and hinders the further exploration of this challenging yet critical problem in the community. In this paper, we propose a large-scale dataset for coronary artery segmentation on CTA images. In addition, we have implemented a benchmark in which we have tried our best to implement several typical existing methods. Furthermore, we propose a strong baseline method which combines multi-scale patch fusion and two-stage processing to extract the details of vessels. Comprehensive experiments show that the proposed method achieves better performance than existing works on the proposed large-scale dataset. The benchmark and the dataset are published at https://github.com/XiaoweiXu/ImageCAS-A-Large-Scale-Dataset-and-Benchmark-for-Coronary-Artery-Segmentation-based-on-CT.
"One-Shot" Reduction of Additive Artifacts in Medical Images
Oct 23, 2021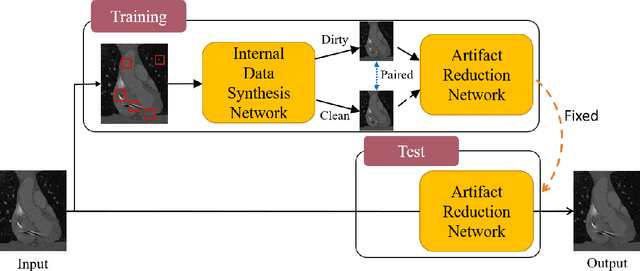
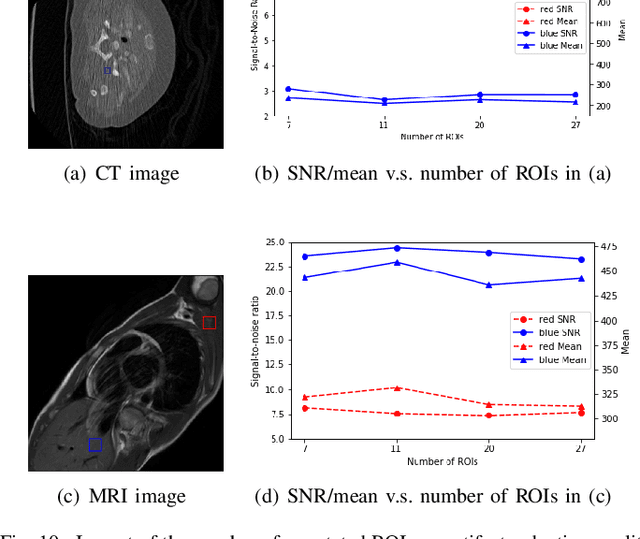
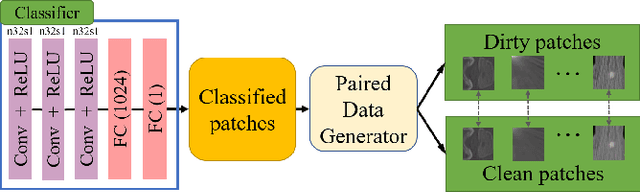
Abstract:Medical images may contain various types of artifacts with different patterns and mixtures, which depend on many factors such as scan setting, machine condition, patients' characteristics, surrounding environment, etc. However, existing deep-learning-based artifact reduction methods are restricted by their training set with specific predetermined artifact types and patterns. As such, they have limited clinical adoption. In this paper, we introduce One-Shot medical image Artifact Reduction (OSAR), which exploits the power of deep learning but without using pre-trained general networks. Specifically, we train a light-weight image-specific artifact reduction network using data synthesized from the input image at test-time. Without requiring any prior large training data set, OSAR can work with almost any medical images that contain varying additive artifacts which are not in any existing data sets. In addition, Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) are used as vehicles and show that the proposed method can reduce artifacts better than state-of-the-art both qualitatively and quantitatively using shorter test time.
Hardware-aware Real-time Myocardial Segmentation Quality Control in Contrast Echocardiography
Sep 14, 2021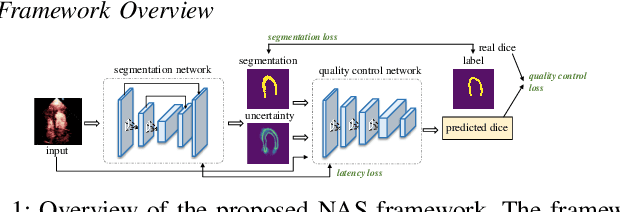
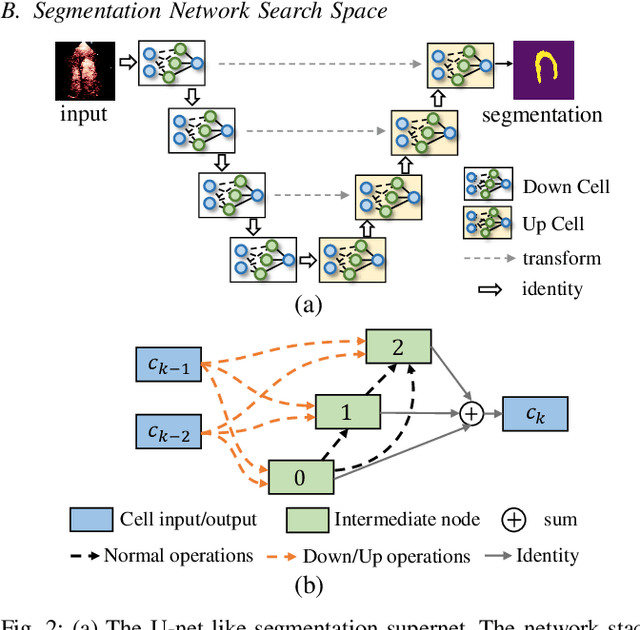
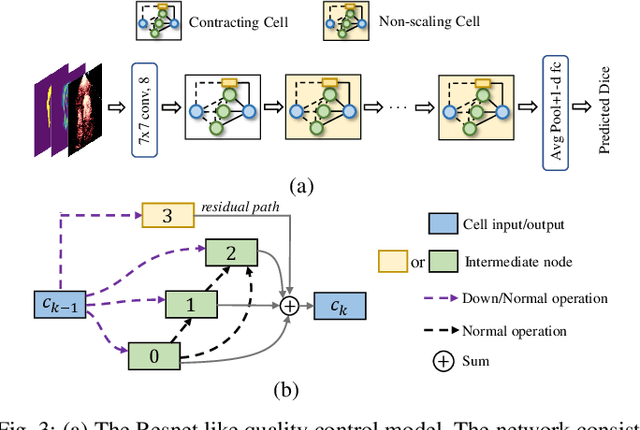
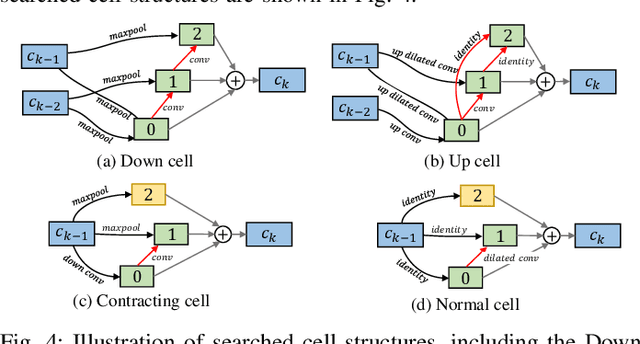
Abstract:Automatic myocardial segmentation of contrast echocardiography has shown great potential in the quantification of myocardial perfusion parameters. Segmentation quality control is an important step to ensure the accuracy of segmentation results for quality research as well as its clinical application. Usually, the segmentation quality control happens after the data acquisition. At the data acquisition time, the operator could not know the quality of the segmentation results. On-the-fly segmentation quality control could help the operator to adjust the ultrasound probe or retake data if the quality is unsatisfied, which can greatly reduce the effort of time-consuming manual correction. However, it is infeasible to deploy state-of-the-art DNN-based models because the segmentation module and quality control module must fit in the limited hardware resource on the ultrasound machine while satisfying strict latency constraints. In this paper, we propose a hardware-aware neural architecture search framework for automatic myocardial segmentation and quality control of contrast echocardiography. We explicitly incorporate the hardware latency as a regularization term into the loss function during training. The proposed method searches the best neural network architecture for the segmentation module and quality prediction module with strict latency.
ImageTBAD: A 3D Computed Tomography Angiography Image Dataset for Automatic Segmentation of Type-B Aortic Dissection
Sep 01, 2021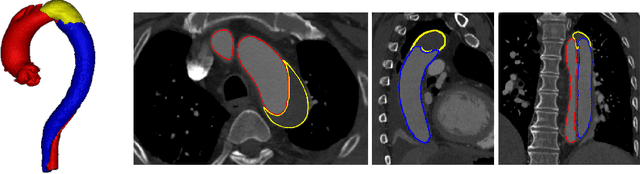

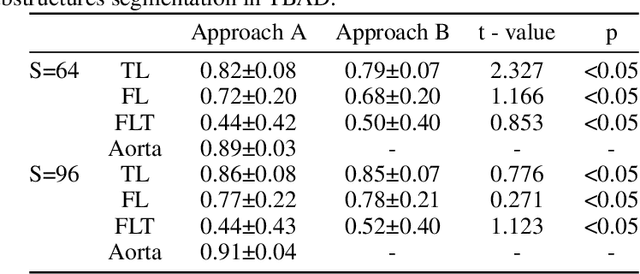
Abstract:Type-B Aortic Dissection (TBAD) is one of the most serious cardiovascular events characterized by a growing yearly incidence,and the severity of disease prognosis. Currently, computed tomography angiography (CTA) has been widely adopted for the diagnosis and prognosis of TBAD. Accurate segmentation of true lumen (TL), false lumen (FL), and false lumen thrombus (FLT) in CTA are crucial for the precise quantification of anatomical features. However, existing works only focus on only TL and FL without considering FLT. In this paper, we propose ImageTBAD, the first 3D computed tomography angiography (CTA) image dataset of TBAD with annotation of TL, FL, and FLT. The proposed dataset contains 100 TBAD CTA images, which is of decent size compared with existing medical imaging datasets. As FLT can appear almost anywhere along the aorta with irregular shapes, segmentation of FLT presents a wide class of segmentation problems where targets exist in a variety of positions with irregular shapes. We further propose a baseline method for automatic segmentation of TBAD. Results show that the baseline method can achieve comparable results with existing works on aorta and TL segmentation. However, the segmentation accuracy of FLT is only 52%, which leaves large room for improvement and also shows the challenge of our dataset. To facilitate further research on this challenging problem, our dataset and codes are released to the public.
Segmentation with Multiple Acceptable Annotations: A Case Study of Myocardial Segmentation in Contrast Echocardiography
Jun 29, 2021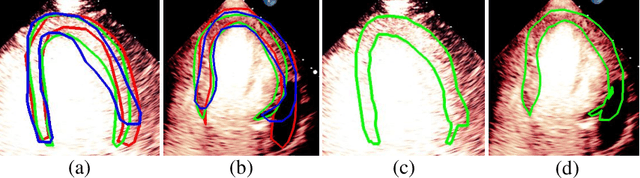
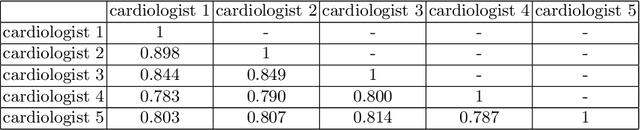
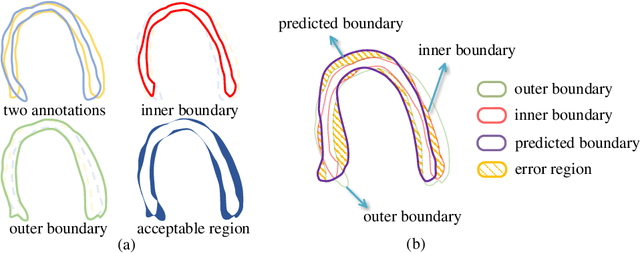
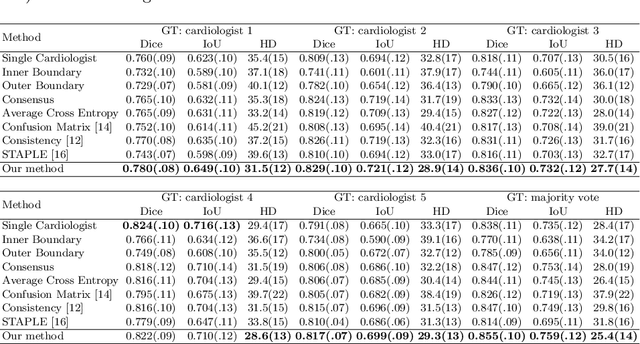
Abstract:Most existing deep learning-based frameworks for image segmentation assume that a unique ground truth is known and can be used for performance evaluation. This is true for many applications, but not all. Myocardial segmentation of Myocardial Contrast Echocardiography (MCE), a critical task in automatic myocardial perfusion analysis, is an example. Due to the low resolution and serious artifacts in MCE data, annotations from different cardiologists can vary significantly, and it is hard to tell which one is the best. In this case, how can we find a good way to evaluate segmentation performance and how do we train the neural network? In this paper, we address the first problem by proposing a new extended Dice to effectively evaluate the segmentation performance when multiple accepted ground truth is available. Then based on our proposed metric, we solve the second problem by further incorporating the new metric into a loss function that enables neural networks to flexibly learn general features of myocardium. Experiment results on our clinical MCE data set demonstrate that the neural network trained with the proposed loss function outperforms those existing ones that try to obtain a unique ground truth from multiple annotations, both quantitatively and qualitatively. Finally, our grading study shows that using extended Dice as an evaluation metric can better identify segmentation results that need manual correction compared with using Dice.
Positional Contrastive Learning for Volumetric Medical Image Segmentation
Jun 18, 2021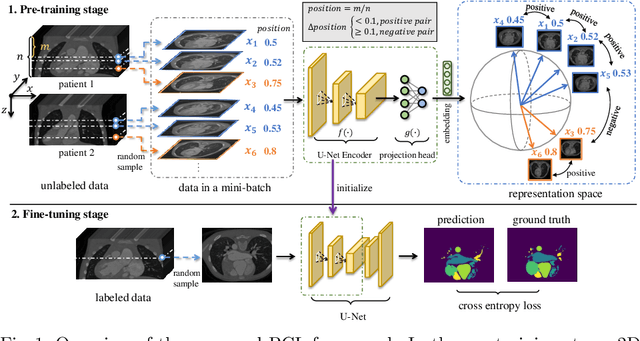
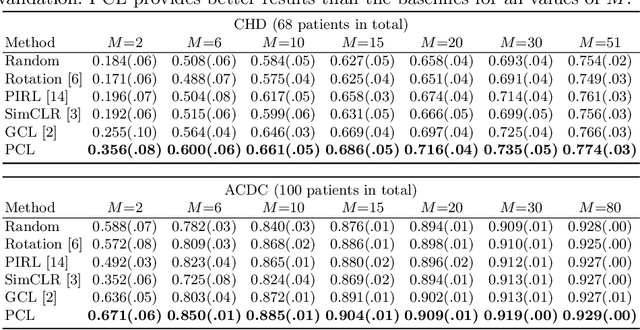
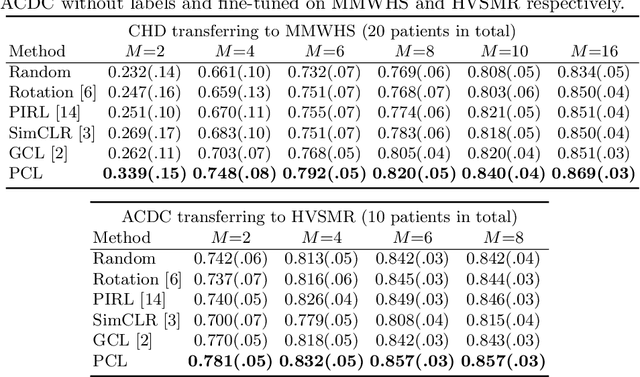
Abstract:The success of deep learning heavily depends on the availability of large labeled training sets. However, it is hard to get large labeled datasets in medical image domain because of the strict privacy concern and costly labeling efforts. Contrastive learning, an unsupervised learning technique, has been proved powerful in learning image-level representations from unlabeled data. The learned encoder can then be transferred or fine-tuned to improve the performance of downstream tasks with limited labels. A critical step in contrastive learning is the generation of contrastive data pairs, which is relatively simple for natural image classification but quite challenging for medical image segmentation due to the existence of the same tissue or organ across the dataset. As a result, when applied to medical image segmentation, most state-of-the-art contrastive learning frameworks inevitably introduce a lot of false-negative pairs and result in degraded segmentation quality. To address this issue, we propose a novel positional contrastive learning (PCL) framework to generate contrastive data pairs by leveraging the position information in volumetric medical images. Experimental results on CT and MRI datasets demonstrate that the proposed PCL method can substantially improve the segmentation performance compared to existing methods in both semi-supervised setting and transfer learning setting.
EchoCP: An Echocardiography Dataset in Contrast Transthoracic Echocardiography for Patent Foramen Ovale Diagnosis
May 18, 2021
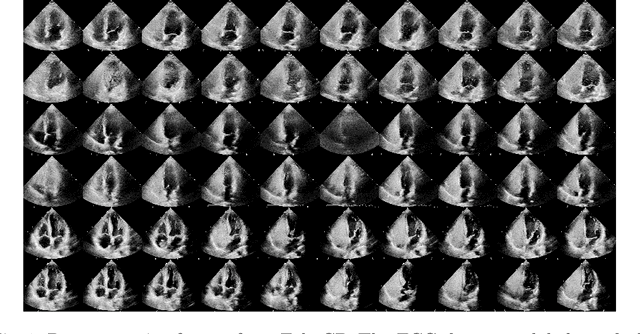
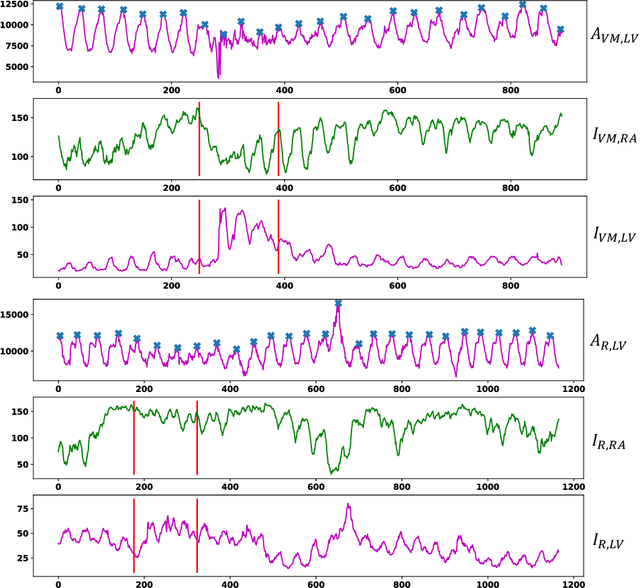
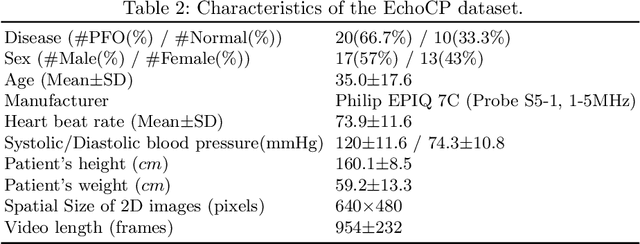
Abstract:Patent foramen ovale (PFO) is a potential separation between the septum, primum and septum secundum located in the anterosuperior portion of the atrial septum. PFO is one of the main factors causing cryptogenic stroke which is the fifth leading cause of death in the United States. For PFO diagnosis, contrast transthoracic echocardiography (cTTE) is preferred as being a more robust method compared with others. However, the current PFO diagnosis through cTTE is extremely slow as it is proceeded manually by sonographers on echocardiography videos. Currently there is no publicly available dataset for this important topic in the community. In this paper, we present EchoCP, as the first echocardiography dataset in cTTE targeting PFO diagnosis. EchoCP consists of 30 patients with both rest and Valsalva maneuver videos which covers various PFO grades. We further establish an automated baseline method for PFO diagnosis based on the state-of-the-art cardiac chamber segmentation technique, which achieves 0.89 average mean Dice score, but only 0.70/0.67 mean accuracies for PFO diagnosis, leaving large room for improvement. We hope that the challenging EchoCP dataset can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released.
Quantization of Deep Neural Networks for Accurate EdgeComputing
Apr 25, 2021
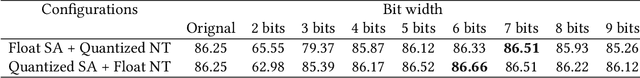
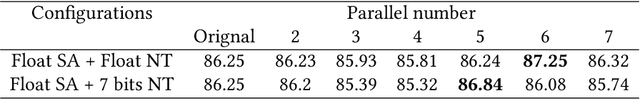
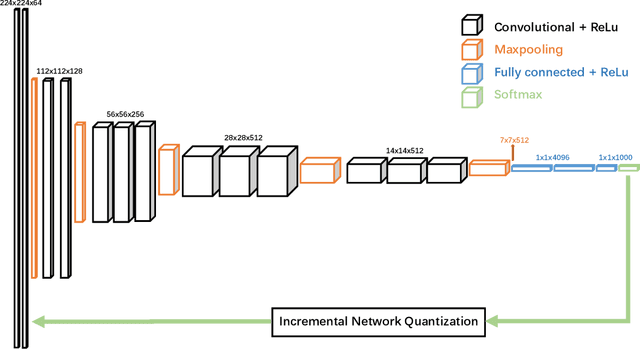
Abstract:Deep neural networks (DNNs) have demonstrated their great potential in recent years, exceeding the per-formance of human experts in a wide range of applications. Due to their large sizes, however, compressiontechniques such as weight quantization and pruning are usually applied before they can be accommodated onthe edge. It is generally believed that quantization leads to performance degradation, and plenty of existingworks have explored quantization strategies aiming at minimum accuracy loss. In this paper, we argue thatquantization, which essentially imposes regularization on weight representations, can sometimes help toimprove accuracy. We conduct comprehensive experiments on three widely used applications: fully con-nected network (FCN) for biomedical image segmentation, convolutional neural network (CNN) for imageclassification on ImageNet, and recurrent neural network (RNN) for automatic speech recognition, and experi-mental results show that quantization can improve the accuracy by 1%, 1.95%, 4.23% on the three applicationsrespectively with 3.5x-6.4x memory reduction.
Multi-Cycle-Consistent Adversarial Networks for Edge Denoising of Computed Tomography Images
Apr 25, 2021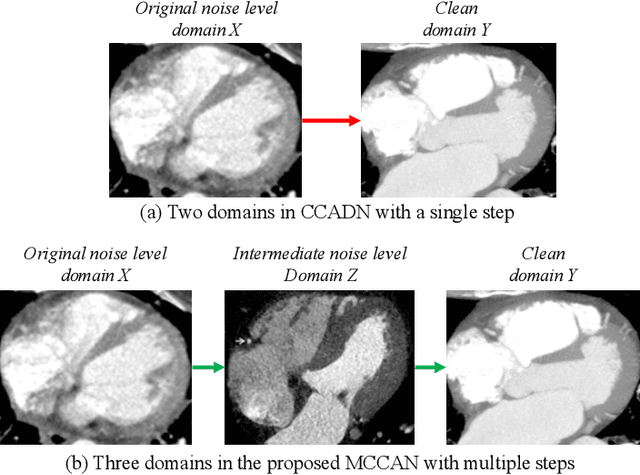
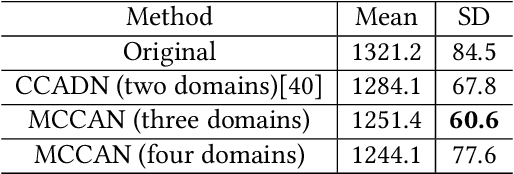
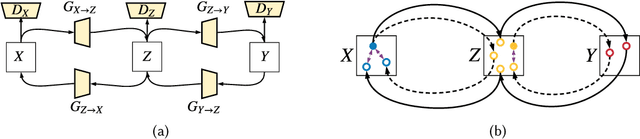
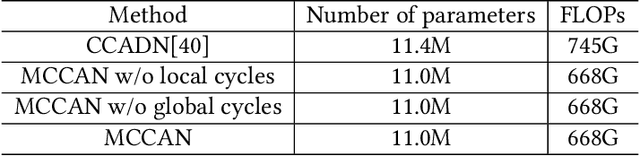
Abstract:As one of the most commonly ordered imaging tests, computed tomography (CT) scan comes with inevitable radiation exposure that increases the cancer risk to patients. However, CT image quality is directly related to radiation dose, thus it is desirable to obtain high-quality CT images with as little dose as possible. CT image denoising tries to obtain high dose like high-quality CT images (domain X) from low dose low-quality CTimages (domain Y), which can be treated as an image-to-image translation task where the goal is to learn the transform between a source domain X (noisy images) and a target domain Y (clean images). In this paper, we propose a multi-cycle-consistent adversarial network (MCCAN) that builds intermediate domains and enforces both local and global cycle-consistency for edge denoising of CT images. The global cycle-consistency couples all generators together to model the whole denoising process, while the local cycle-consistency imposes effective supervision on the process between adjacent domains. Experiments show that both local and global cycle-consistency are important for the success of MCCAN, which outperformsCCADN in terms of denoising quality with slightly less computation resource consumption.
ImageCHD: A 3D Computed Tomography Image Dataset for Classification of Congenital Heart Disease
Jan 26, 2021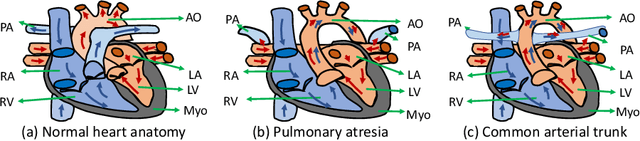
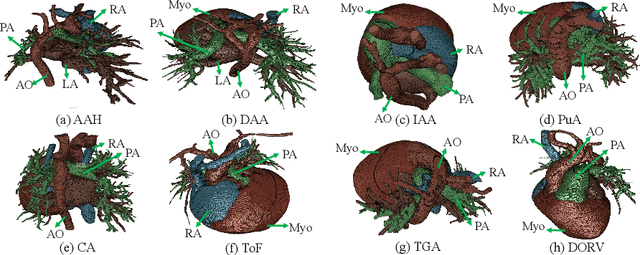
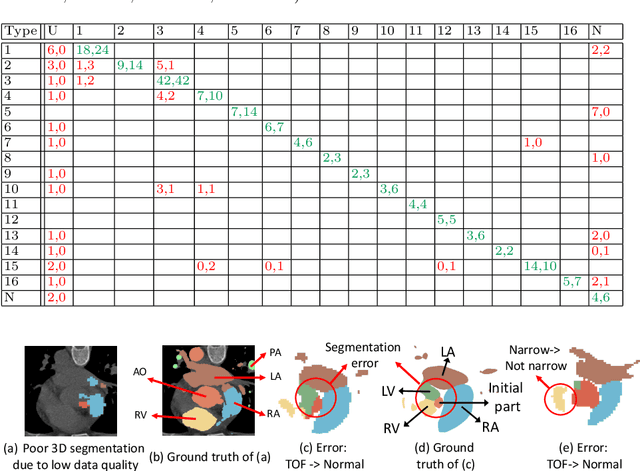
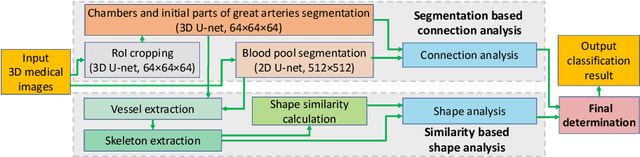
Abstract:Congenital heart disease (CHD) is the most common type of birth defect, which occurs 1 in every 110 births in the United States. CHD usually comes with severe variations in heart structure and great artery connections that can be classified into many types. Thus highly specialized domain knowledge and the time-consuming human process is needed to analyze the associated medical images. On the other hand, due to the complexity of CHD and the lack of dataset, little has been explored on the automatic diagnosis (classification) of CHDs. In this paper, we present ImageCHD, the first medical image dataset for CHD classification. ImageCHD contains 110 3D Computed Tomography (CT) images covering most types of CHD, which is of decent size Classification of CHDs requires the identification of large structural changes without any local tissue changes, with limited data. It is an example of a larger class of problems that are quite difficult for current machine-learning-based vision methods to solve. To demonstrate this, we further present a baseline framework for the automatic classification of CHD, based on a state-of-the-art CHD segmentation method. Experimental results show that the baseline framework can only achieve a classification accuracy of 82.0\% under a selective prediction scheme with 88.4\% coverage, leaving big room for further improvement. We hope that ImageCHD can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released to the public compared with existing medical imaging datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge