Dewen Zeng
Contrastive Learning with Synthetic Positives
Aug 30, 2024Abstract:Contrastive learning with the nearest neighbor has proved to be one of the most efficient self-supervised learning (SSL) techniques by utilizing the similarity of multiple instances within the same class. However, its efficacy is constrained as the nearest neighbor algorithm primarily identifies ``easy'' positive pairs, where the representations are already closely located in the embedding space. In this paper, we introduce a novel approach called Contrastive Learning with Synthetic Positives (CLSP) that utilizes synthetic images, generated by an unconditional diffusion model, as the additional positives to help the model learn from diverse positives. Through feature interpolation in the diffusion model sampling process, we generate images with distinct backgrounds yet similar semantic content to the anchor image. These images are considered ``hard'' positives for the anchor image, and when included as supplementary positives in the contrastive loss, they contribute to a performance improvement of over 2\% and 1\% in linear evaluation compared to the previous NNCLR and All4One methods across multiple benchmark datasets such as CIFAR10, achieving state-of-the-art methods. On transfer learning benchmarks, CLSP outperforms existing SSL frameworks on 6 out of 8 downstream datasets. We believe CLSP establishes a valuable baseline for future SSL studies incorporating synthetic data in the training process.
Enhancing 3D Transformer Segmentation Model for Medical Image with Token-level Representation Learning
Aug 12, 2024Abstract:In the field of medical images, although various works find Swin Transformer has promising effectiveness on pixelwise dense prediction, whether pre-training these models without using extra dataset can further boost the performance for the downstream semantic segmentation remains unexplored.Applications of previous representation learning methods are hindered by the limited number of 3D volumes and high computational cost. In addition, most of pretext tasks designed specifically for Transformer are not applicable to hierarchical structure of Swin Transformer. Thus, this work proposes a token-level representation learning loss that maximizes agreement between token embeddings from different augmented views individually instead of volume-level global features. Moreover, we identify a potential representation collapse exclusively caused by this new loss. To prevent collapse, we invent a simple "rotate-and-restore" mechanism, which rotates and flips one augmented view of input volume, and later restores the order of tokens in the feature maps. We also modify the contrastive loss to address the discrimination between tokens at the same position but from different volumes. We test our pre-training scheme on two public medical segmentation datasets, and the results on the downstream segmentation task show more improvement of our methods than other state-of-the-art pre-trainig methods.
Achieving Fairness Through Channel Pruning for Dermatological Disease Diagnosis
May 14, 2024



Abstract:Numerous studies have revealed that deep learning-based medical image classification models may exhibit bias towards specific demographic attributes, such as race, gender, and age. Existing bias mitigation methods often achieve high level of fairness at the cost of significant accuracy degradation. In response to this challenge, we propose an innovative and adaptable Soft Nearest Neighbor Loss-based channel pruning framework, which achieves fairness through channel pruning. Traditionally, channel pruning is utilized to accelerate neural network inference. However, our work demonstrates that pruning can also be a potent tool for achieving fairness. Our key insight is that different channels in a layer contribute differently to the accuracy of different groups. By selectively pruning critical channels that lead to the accuracy difference between the privileged and unprivileged groups, we can effectively improve fairness without sacrificing accuracy significantly. Experiments conducted on two skin lesion diagnosis datasets across multiple sensitive attributes validate the effectiveness of our method in achieving state-of-the-art trade-off between accuracy and fairness. Our code is available at https://github.com/Kqp1227/Sensitive-Channel-Pruning.
Robust Implementation of Retrieval-Augmented Generation on Edge-based Computing-in-Memory Architectures
May 07, 2024



Abstract:Large Language Models (LLMs) deployed on edge devices learn through fine-tuning and updating a certain portion of their parameters. Although such learning methods can be optimized to reduce resource utilization, the overall required resources remain a heavy burden on edge devices. Instead, Retrieval-Augmented Generation (RAG), a resource-efficient LLM learning method, can improve the quality of the LLM-generated content without updating model parameters. However, the RAG-based LLM may involve repetitive searches on the profile data in every user-LLM interaction. This search can lead to significant latency along with the accumulation of user data. Conventional efforts to decrease latency result in restricting the size of saved user data, thus reducing the scalability of RAG as user data continuously grows. It remains an open question: how to free RAG from the constraints of latency and scalability on edge devices? In this paper, we propose a novel framework to accelerate RAG via Computing-in-Memory (CiM) architectures. It accelerates matrix multiplications by performing in-situ computation inside the memory while avoiding the expensive data transfer between the computing unit and memory. Our framework, Robust CiM-backed RAG (RoCR), utilizing a novel contrastive learning-based training method and noise-aware training, can enable RAG to efficiently search profile data with CiM. To the best of our knowledge, this is the first work utilizing CiM to accelerate RAG.
Learning to Skip for Language Modeling
Nov 26, 2023



Abstract:Overparameterized large-scale language models have impressive generalization performance of in-context few-shot learning. However, most language models allocate the same amount of parameters or computation to each token, disregarding the complexity or importance of the input data. We argue that in language model pretraining, a variable amount of computation should be assigned to different tokens, and this can be efficiently achieved via a simple routing mechanism. Different from conventional early stopping techniques where tokens can early exit at only early layers, we propose a more general method that dynamically skips the execution of a layer (or module) for any input token with a binary router. In our extensive evaluation across 24 NLP tasks, we demonstrate that the proposed method can significantly improve the 1-shot performance compared to other competitive baselines only at mild extra cost for inference.
Additional Positive Enables Better Representation Learning for Medical Images
May 31, 2023


Abstract:This paper presents a new way to identify additional positive pairs for BYOL, a state-of-the-art (SOTA) self-supervised learning framework, to improve its representation learning ability. Unlike conventional BYOL which relies on only one positive pair generated by two augmented views of the same image, we argue that information from different images with the same label can bring more diversity and variations to the target features, thus benefiting representation learning. To identify such pairs without any label, we investigate TracIn, an instance-based and computationally efficient influence function, for BYOL training. Specifically, TracIn is a gradient-based method that reveals the impact of a training sample on a test sample in supervised learning. We extend it to the self-supervised learning setting and propose an efficient batch-wise per-sample gradient computation method to estimate the pairwise TracIn to represent the similarity of samples in the mini-batch during training. For each image, we select the most similar sample from other images as the additional positive and pull their features together with BYOL loss. Experimental results on two public medical datasets (i.e., ISIC 2019 and ChestX-ray) demonstrate that the proposed method can improve the classification performance compared to other competitive baselines in both semi-supervised and transfer learning settings.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
FedCoCo: A Memory Efficient Federated Self-supervised Framework for On-Device Visual Representation Learning
Dec 02, 2022



Abstract:The ubiquity of edge devices has led to a growing amount of unlabeled data produced at the edge. Deep learning models deployed on edge devices are required to learn from these unlabeled data to continuously improve accuracy. Self-supervised representation learning has achieved promising performances using centralized unlabeled data. However, the increasing awareness of privacy protection limits centralizing the distributed unlabeled image data on edge devices. While federated learning has been widely adopted to enable distributed machine learning with privacy preservation, without a data selection method to efficiently select streaming data, the traditional federated learning framework fails to handle these huge amounts of decentralized unlabeled data with limited storage resources on edge. To address these challenges, we propose a Federated on-device Contrastive learning framework with Coreset selection, which we call FedCoCo, to automatically select a coreset that consists of the most representative samples into the replay buffer on each device. It preserves data privacy as each client does not share raw data while learning good visual representations. Experiments demonstrate the effectiveness and significance of the proposed method in visual representation learning.
Federated Self-Supervised Contrastive Learning and Masked Autoencoder for Dermatological Disease Diagnosis
Aug 24, 2022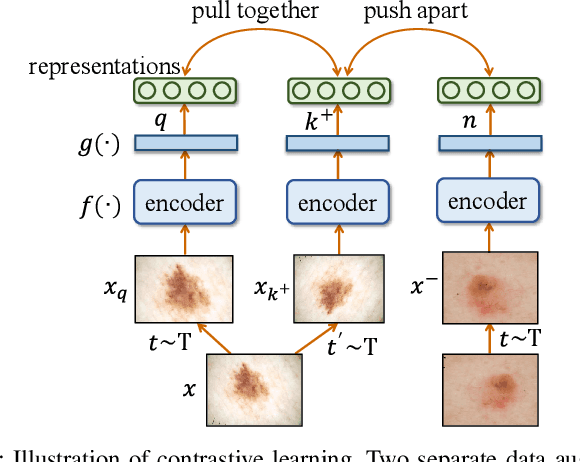
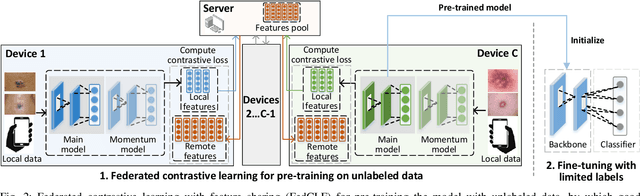
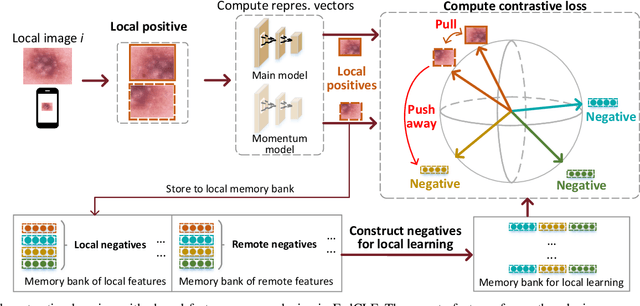
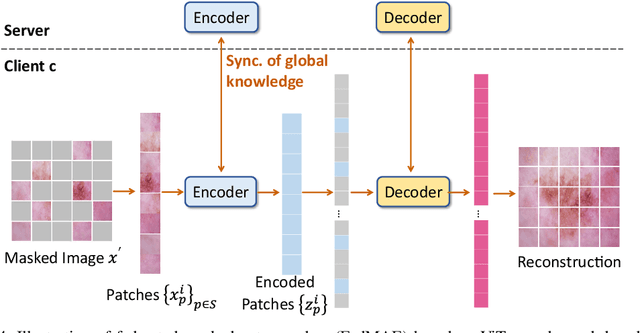
Abstract:In dermatological disease diagnosis, the private data collected by mobile dermatology assistants exist on distributed mobile devices of patients. Federated learning (FL) can use decentralized data to train models while keeping data local. Existing FL methods assume all the data have labels. However, medical data often comes without full labels due to high labeling costs. Self-supervised learning (SSL) methods, contrastive learning (CL) and masked autoencoders (MAE), can leverage the unlabeled data to pre-train models, followed by fine-tuning with limited labels. However, combining SSL and FL has unique challenges. For example, CL requires diverse data but each device only has limited data. For MAE, while Vision Transformer (ViT) based MAE has higher accuracy over CNNs in centralized learning, MAE's performance in FL with unlabeled data has not been investigated. Besides, the ViT synchronization between the server and clients is different from traditional CNNs. Therefore, special synchronization methods need to be designed. In this work, we propose two federated self-supervised learning frameworks for dermatological disease diagnosis with limited labels. The first one features lower computation costs, suitable for mobile devices. The second one features high accuracy and fits high-performance servers. Based on CL, we proposed federated contrastive learning with feature sharing (FedCLF). Features are shared for diverse contrastive information without sharing raw data for privacy. Based on MAE, we proposed FedMAE. Knowledge split separates the global and local knowledge learned from each client. Only global knowledge is aggregated for higher generalization performance. Experiments on dermatological disease datasets show superior accuracy of the proposed frameworks over state-of-the-arts.
Distributed Contrastive Learning for Medical Image Segmentation
Aug 07, 2022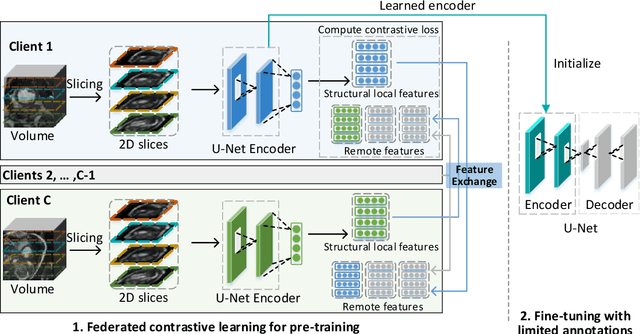
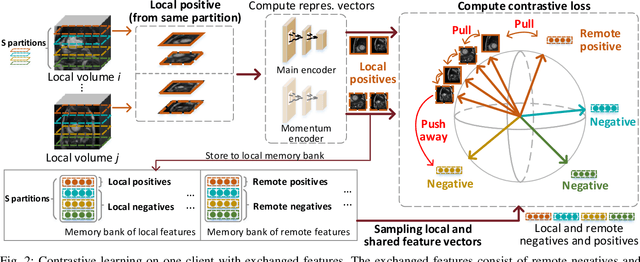
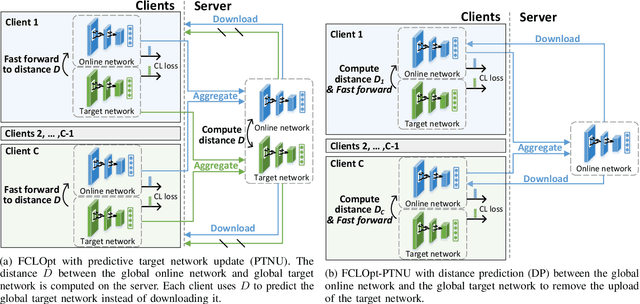
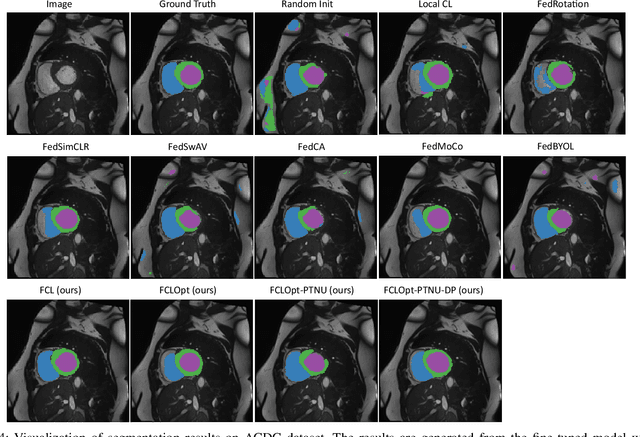
Abstract:Supervised deep learning needs a large amount of labeled data to achieve high performance. However, in medical imaging analysis, each site may only have a limited amount of data and labels, which makes learning ineffective. Federated learning (FL) can learn a shared model from decentralized data. But traditional FL requires fully-labeled data for training, which is very expensive to obtain. Self-supervised contrastive learning (CL) can learn from unlabeled data for pre-training, followed by fine-tuning with limited annotations. However, when adopting CL in FL, the limited data diversity on each site makes federated contrastive learning (FCL) ineffective. In this work, we propose two federated self-supervised learning frameworks for volumetric medical image segmentation with limited annotations. The first one features high accuracy and fits high-performance servers with high-speed connections. The second one features lower communication costs, suitable for mobile devices. In the first framework, features are exchanged during FCL to provide diverse contrastive data to each site for effective local CL while keeping raw data private. Global structural matching aligns local and remote features for a unified feature space among different sites. In the second framework, to reduce the communication cost for feature exchanging, we propose an optimized method FCLOpt that does not rely on negative samples. To reduce the communications of model download, we propose the predictive target network update (PTNU) that predicts the parameters of the target network. Based on PTNU, we propose the distance prediction (DP) to remove most of the uploads of the target network. Experiments on a cardiac MRI dataset show the proposed two frameworks substantially improve the segmentation and generalization performance compared with state-of-the-art techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge