Xiaowei Xu
Multi-Layer Pseudo-Supervision for Histopathology Tissue Semantic Segmentation using Patch-level Classification Labels
Oct 14, 2021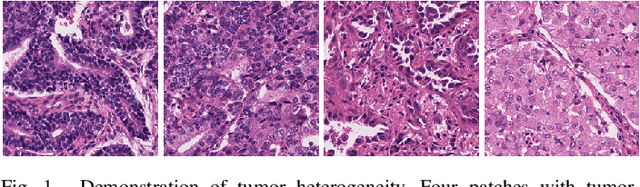

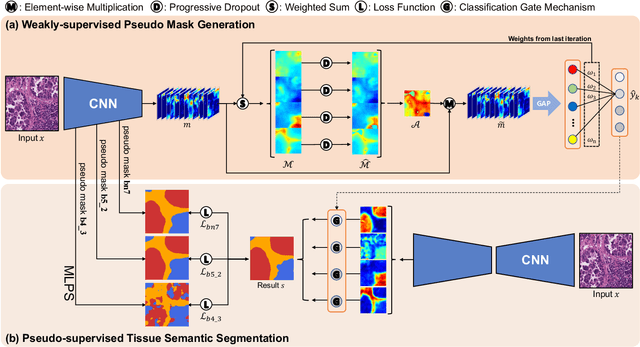
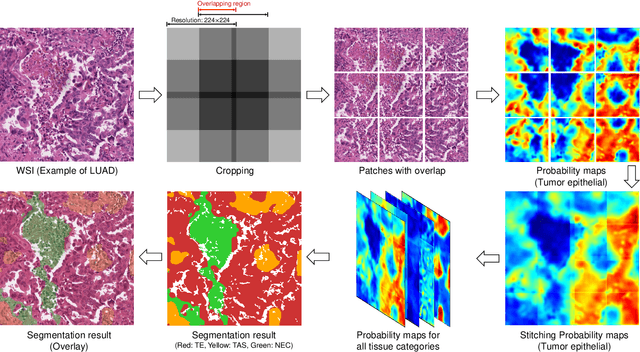
Abstract:Tissue-level semantic segmentation is a vital step in computational pathology. Fully-supervised models have already achieved outstanding performance with dense pixel-level annotations. However, drawing such labels on the giga-pixel whole slide images is extremely expensive and time-consuming. In this paper, we use only patch-level classification labels to achieve tissue semantic segmentation on histopathology images, finally reducing the annotation efforts. We proposed a two-step model including a classification and a segmentation phases. In the classification phase, we proposed a CAM-based model to generate pseudo masks by patch-level labels. In the segmentation phase, we achieved tissue semantic segmentation by our proposed Multi-Layer Pseudo-Supervision. Several technical novelties have been proposed to reduce the information gap between pixel-level and patch-level annotations. As a part of this paper, we introduced a new weakly-supervised semantic segmentation (WSSS) dataset for lung adenocarcinoma (LUAD-HistoSeg). We conducted several experiments to evaluate our proposed model on two datasets. Our proposed model outperforms two state-of-the-art WSSS approaches. Note that we can achieve comparable quantitative and qualitative results with the fully-supervised model, with only around a 2\% gap for MIoU and FwIoU. By comparing with manual labeling, our model can greatly save the annotation time from hours to minutes. The source code is available at: \url{https://github.com/ChuHan89/WSSS-Tissue}.
Semi-supervised Contrastive Learning for Label-efficient Medical Image Segmentation
Sep 28, 2021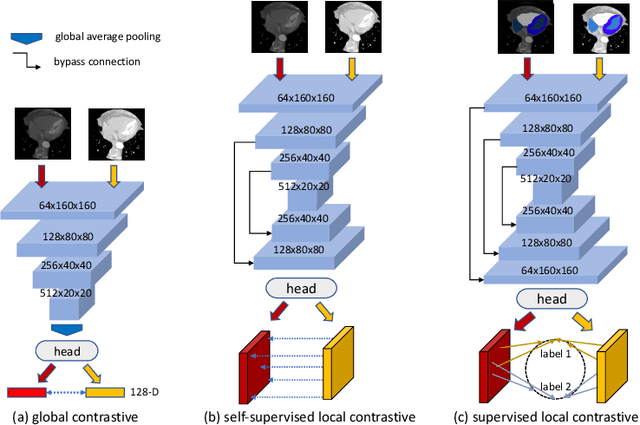
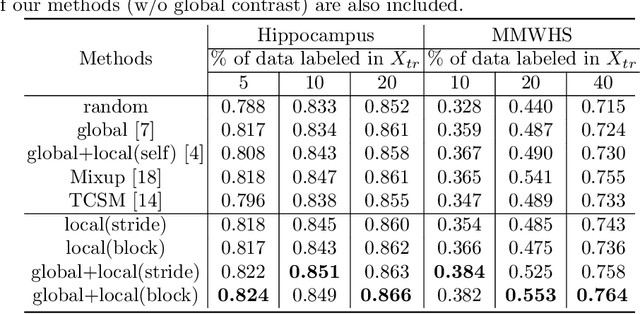
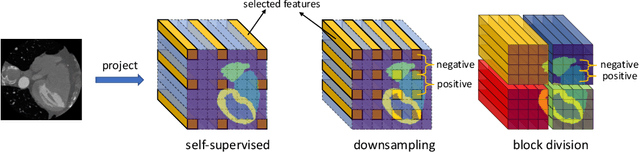
Abstract:The success of deep learning methods in medical image segmentation tasks heavily depends on a large amount of labeled data to supervise the training. On the other hand, the annotation of biomedical images requires domain knowledge and can be laborious. Recently, contrastive learning has demonstrated great potential in learning latent representation of images even without any label. Existing works have explored its application to biomedical image segmentation where only a small portion of data is labeled, through a pre-training phase based on self-supervised contrastive learning without using any labels followed by a supervised fine-tuning phase on the labeled portion of data only. In this paper, we establish that by including the limited label in formation in the pre-training phase, it is possible to boost the performance of contrastive learning. We propose a supervised local contrastive loss that leverages limited pixel-wise annotation to force pixels with the same label to gather around in the embedding space. Such loss needs pixel-wise computation which can be expensive for large images, and we further propose two strategies, downsampling and block division, to address the issue. We evaluate our methods on two public biomedical image datasets of different modalities. With different amounts of labeled data, our methods consistently outperform the state-of-the-art contrast-based methods and other semi-supervised learning techniques.
Hardware-aware Real-time Myocardial Segmentation Quality Control in Contrast Echocardiography
Sep 14, 2021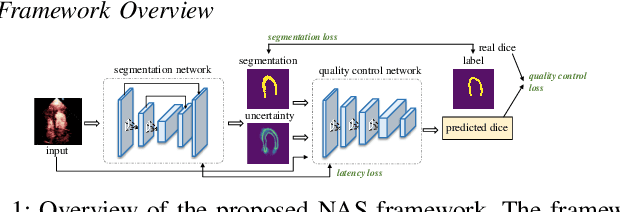
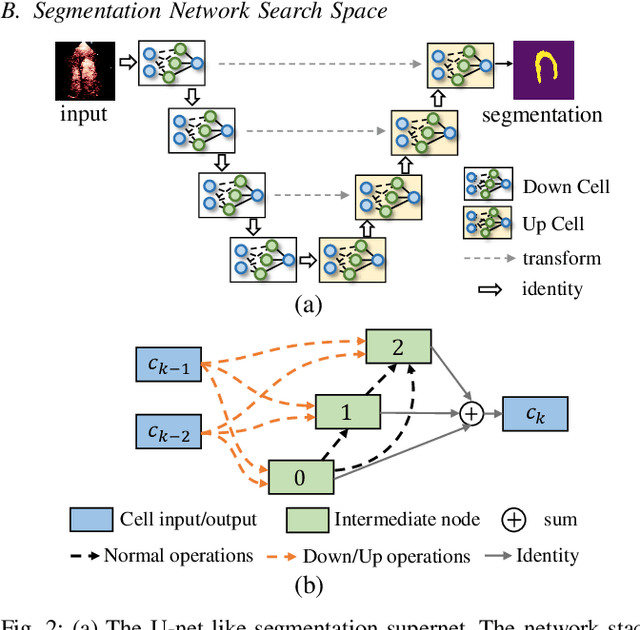
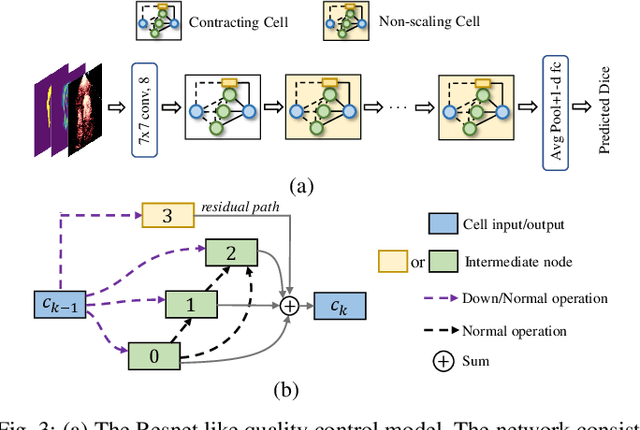
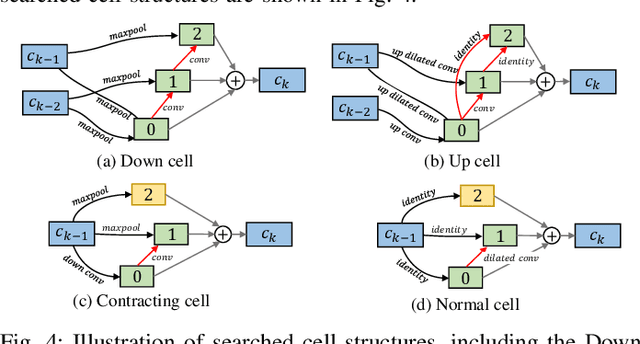
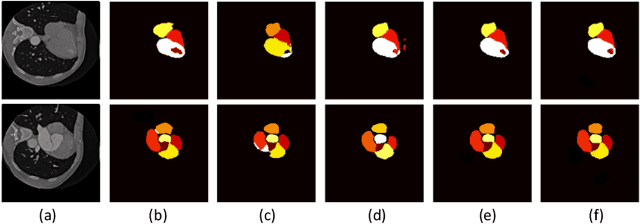
Abstract:Automatic myocardial segmentation of contrast echocardiography has shown great potential in the quantification of myocardial perfusion parameters. Segmentation quality control is an important step to ensure the accuracy of segmentation results for quality research as well as its clinical application. Usually, the segmentation quality control happens after the data acquisition. At the data acquisition time, the operator could not know the quality of the segmentation results. On-the-fly segmentation quality control could help the operator to adjust the ultrasound probe or retake data if the quality is unsatisfied, which can greatly reduce the effort of time-consuming manual correction. However, it is infeasible to deploy state-of-the-art DNN-based models because the segmentation module and quality control module must fit in the limited hardware resource on the ultrasound machine while satisfying strict latency constraints. In this paper, we propose a hardware-aware neural architecture search framework for automatic myocardial segmentation and quality control of contrast echocardiography. We explicitly incorporate the hardware latency as a regularization term into the loss function during training. The proposed method searches the best neural network architecture for the segmentation module and quality prediction module with strict latency.
ImageTBAD: A 3D Computed Tomography Angiography Image Dataset for Automatic Segmentation of Type-B Aortic Dissection
Sep 01, 2021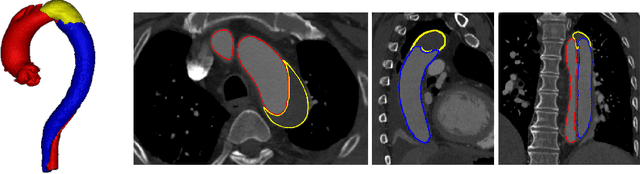
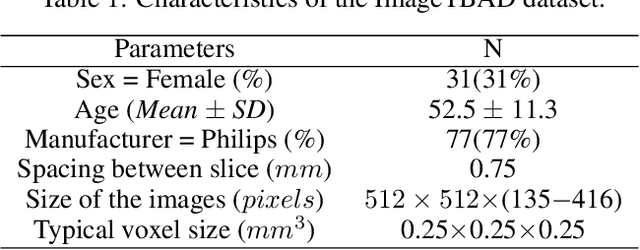

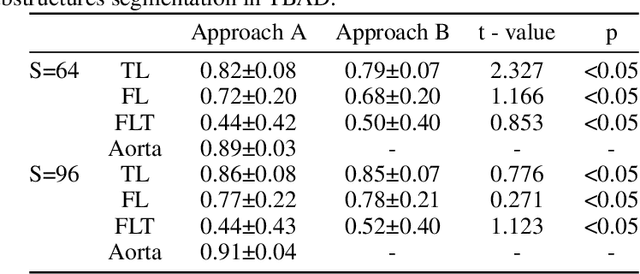
Abstract:Type-B Aortic Dissection (TBAD) is one of the most serious cardiovascular events characterized by a growing yearly incidence,and the severity of disease prognosis. Currently, computed tomography angiography (CTA) has been widely adopted for the diagnosis and prognosis of TBAD. Accurate segmentation of true lumen (TL), false lumen (FL), and false lumen thrombus (FLT) in CTA are crucial for the precise quantification of anatomical features. However, existing works only focus on only TL and FL without considering FLT. In this paper, we propose ImageTBAD, the first 3D computed tomography angiography (CTA) image dataset of TBAD with annotation of TL, FL, and FLT. The proposed dataset contains 100 TBAD CTA images, which is of decent size compared with existing medical imaging datasets. As FLT can appear almost anywhere along the aorta with irregular shapes, segmentation of FLT presents a wide class of segmentation problems where targets exist in a variety of positions with irregular shapes. We further propose a baseline method for automatic segmentation of TBAD. Results show that the baseline method can achieve comparable results with existing works on aorta and TL segmentation. However, the segmentation accuracy of FLT is only 52%, which leaves large room for improvement and also shows the challenge of our dataset. To facilitate further research on this challenging problem, our dataset and codes are released to the public.
Segmentation with Multiple Acceptable Annotations: A Case Study of Myocardial Segmentation in Contrast Echocardiography
Jun 29, 2021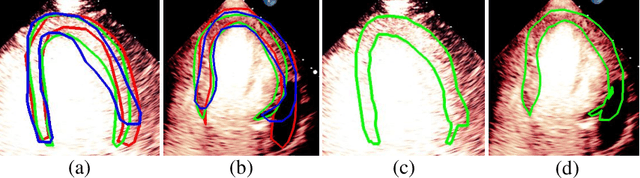
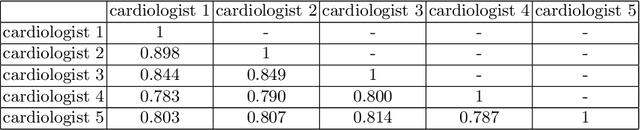
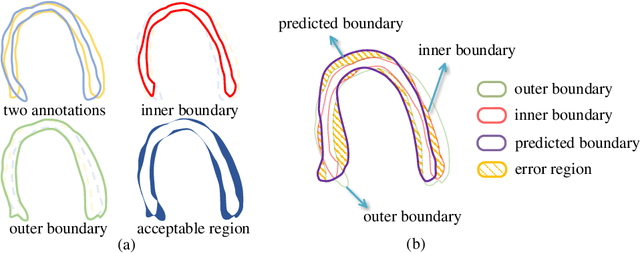
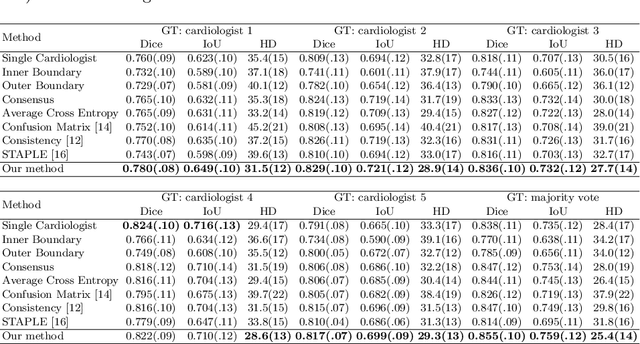
Abstract:Most existing deep learning-based frameworks for image segmentation assume that a unique ground truth is known and can be used for performance evaluation. This is true for many applications, but not all. Myocardial segmentation of Myocardial Contrast Echocardiography (MCE), a critical task in automatic myocardial perfusion analysis, is an example. Due to the low resolution and serious artifacts in MCE data, annotations from different cardiologists can vary significantly, and it is hard to tell which one is the best. In this case, how can we find a good way to evaluate segmentation performance and how do we train the neural network? In this paper, we address the first problem by proposing a new extended Dice to effectively evaluate the segmentation performance when multiple accepted ground truth is available. Then based on our proposed metric, we solve the second problem by further incorporating the new metric into a loss function that enables neural networks to flexibly learn general features of myocardium. Experiment results on our clinical MCE data set demonstrate that the neural network trained with the proposed loss function outperforms those existing ones that try to obtain a unique ground truth from multiple annotations, both quantitatively and qualitatively. Finally, our grading study shows that using extended Dice as an evaluation metric can better identify segmentation results that need manual correction compared with using Dice.
Positional Contrastive Learning for Volumetric Medical Image Segmentation
Jun 18, 2021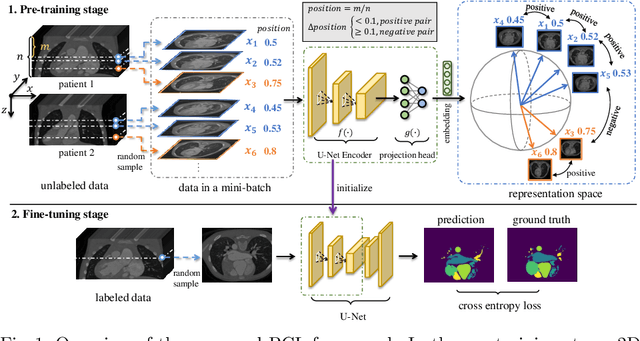
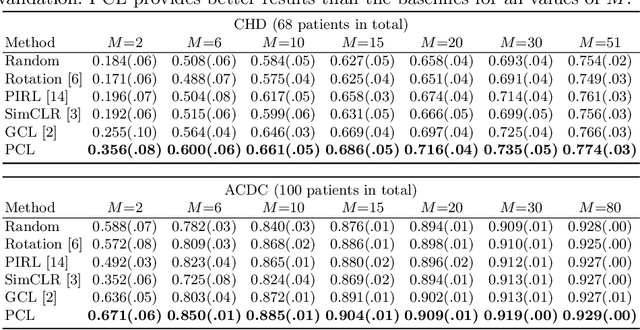
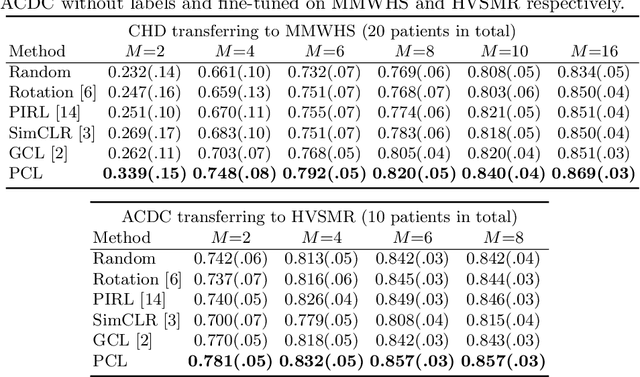
Abstract:The success of deep learning heavily depends on the availability of large labeled training sets. However, it is hard to get large labeled datasets in medical image domain because of the strict privacy concern and costly labeling efforts. Contrastive learning, an unsupervised learning technique, has been proved powerful in learning image-level representations from unlabeled data. The learned encoder can then be transferred or fine-tuned to improve the performance of downstream tasks with limited labels. A critical step in contrastive learning is the generation of contrastive data pairs, which is relatively simple for natural image classification but quite challenging for medical image segmentation due to the existence of the same tissue or organ across the dataset. As a result, when applied to medical image segmentation, most state-of-the-art contrastive learning frameworks inevitably introduce a lot of false-negative pairs and result in degraded segmentation quality. To address this issue, we propose a novel positional contrastive learning (PCL) framework to generate contrastive data pairs by leveraging the position information in volumetric medical images. Experimental results on CT and MRI datasets demonstrate that the proposed PCL method can substantially improve the segmentation performance compared to existing methods in both semi-supervised setting and transfer learning setting.
EchoCP: An Echocardiography Dataset in Contrast Transthoracic Echocardiography for Patent Foramen Ovale Diagnosis
May 18, 2021
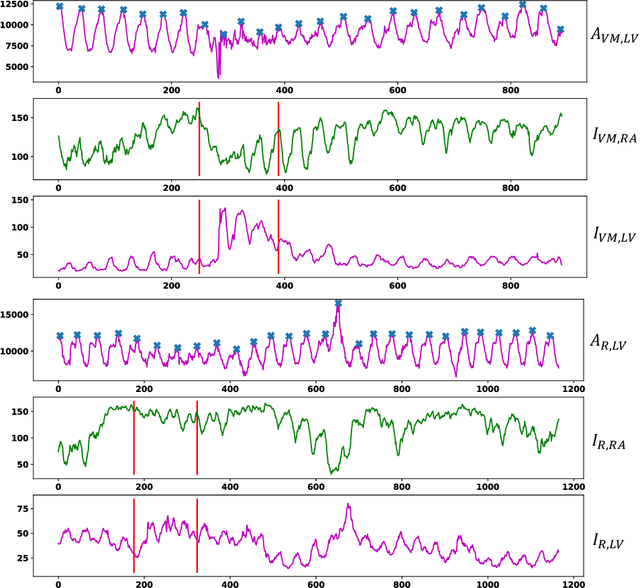
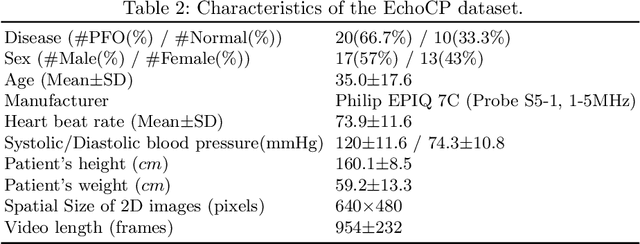
Abstract:Patent foramen ovale (PFO) is a potential separation between the septum, primum and septum secundum located in the anterosuperior portion of the atrial septum. PFO is one of the main factors causing cryptogenic stroke which is the fifth leading cause of death in the United States. For PFO diagnosis, contrast transthoracic echocardiography (cTTE) is preferred as being a more robust method compared with others. However, the current PFO diagnosis through cTTE is extremely slow as it is proceeded manually by sonographers on echocardiography videos. Currently there is no publicly available dataset for this important topic in the community. In this paper, we present EchoCP, as the first echocardiography dataset in cTTE targeting PFO diagnosis. EchoCP consists of 30 patients with both rest and Valsalva maneuver videos which covers various PFO grades. We further establish an automated baseline method for PFO diagnosis based on the state-of-the-art cardiac chamber segmentation technique, which achieves 0.89 average mean Dice score, but only 0.70/0.67 mean accuracies for PFO diagnosis, leaving large room for improvement. We hope that the challenging EchoCP dataset can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released.
Pyramid U-Net for Retinal Vessel Segmentation
Apr 06, 2021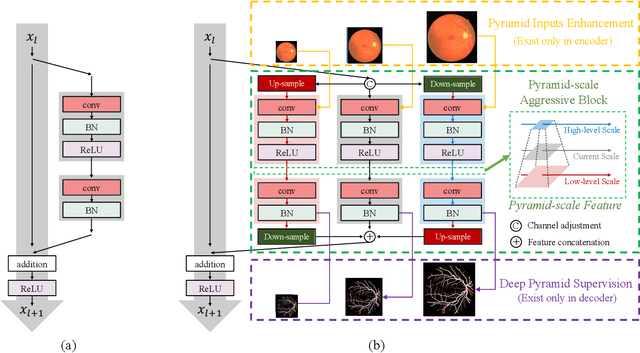
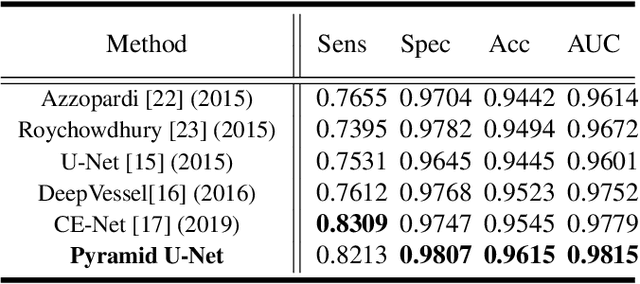
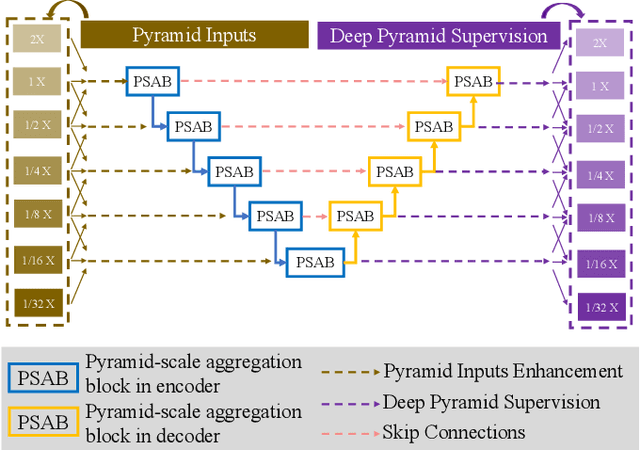

Abstract:Retinal blood vessel can assist doctors in diagnosis of eye-related diseases such as diabetes and hypertension, and its segmentation is particularly important for automatic retinal image analysis. However, it is challenging to segment these vessels structures, especially the thin capillaries from the color retinal image due to low contrast and ambiguousness. In this paper, we propose pyramid U-Net for accurate retinal vessel segmentation. In pyramid U-Net, the proposed pyramid-scale aggregation blocks (PSABs) are employed in both the encoder and decoder to aggregate features at higher, current and lower levels. In this way, coarse-to-fine context information is shared and aggregated in each block thus to improve the location of capillaries. To further improve performance, two optimizations including pyramid inputs enhancement and deep pyramid supervision are applied to PSABs in the encoder and decoder, respectively. For PSABs in the encoder, scaled input images are added as extra inputs. While for PSABs in the decoder, scaled intermediate outputs are supervised by the scaled segmentation labels. Extensive evaluations show that our pyramid U-Net outperforms the current state-of-the-art methods on the public DRIVE and CHASE-DB1 datasets.
ObjectAug: Object-level Data Augmentation for Semantic Image Segmentation
Jan 30, 2021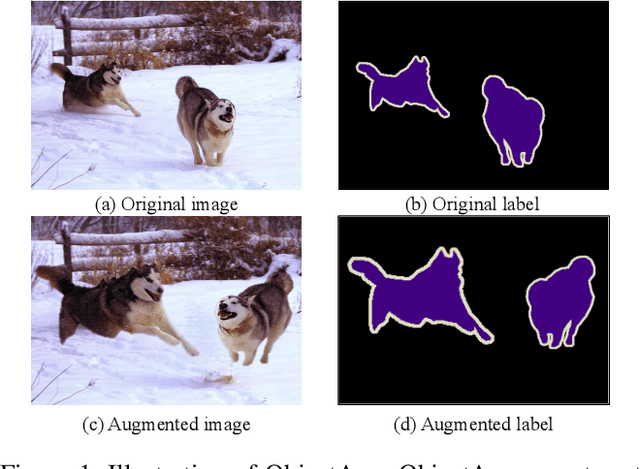
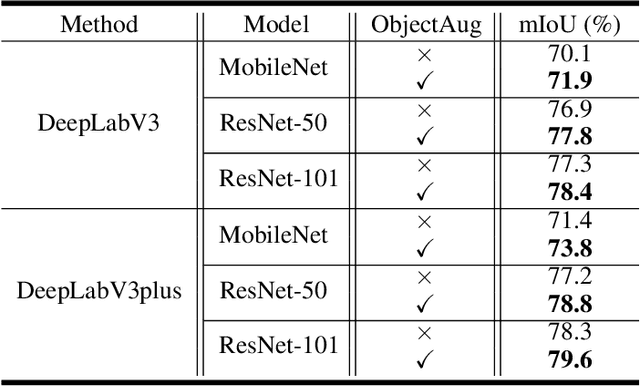
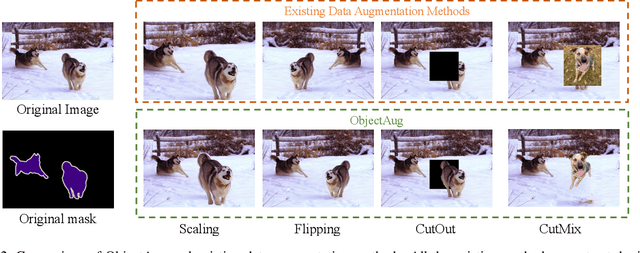
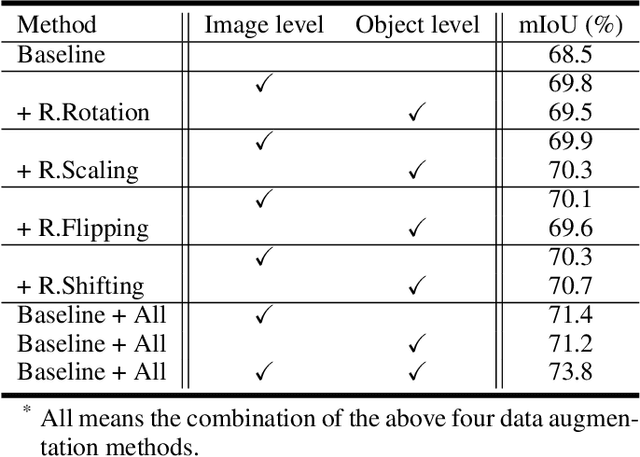
Abstract:Semantic image segmentation aims to obtain object labels with precise boundaries, which usually suffers from overfitting. Recently, various data augmentation strategies like regional dropout and mix strategies have been proposed to address the problem. These strategies have proved to be effective for guiding the model to attend on less discriminative parts. However, current strategies operate at the image level, and objects and the background are coupled. Thus, the boundaries are not well augmented due to the fixed semantic scenario. In this paper, we propose ObjectAug to perform object-level augmentation for semantic image segmentation. ObjectAug first decouples the image into individual objects and the background using the semantic labels. Next, each object is augmented individually with commonly used augmentation methods (e.g., scaling, shifting, and rotation). Then, the black area brought by object augmentation is further restored using image inpainting. Finally, the augmented objects and background are assembled as an augmented image. In this way, the boundaries can be fully explored in the various semantic scenarios. In addition, ObjectAug can support category-aware augmentation that gives various possibilities to objects in each category, and can be easily combined with existing image-level augmentation methods to further boost performance. Comprehensive experiments are conducted on both natural image and medical image datasets. Experiment results demonstrate that our ObjectAug can evidently improve segmentation performance.
ImageCHD: A 3D Computed Tomography Image Dataset for Classification of Congenital Heart Disease
Jan 26, 2021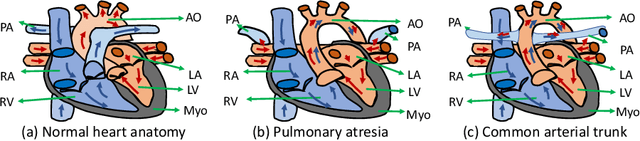
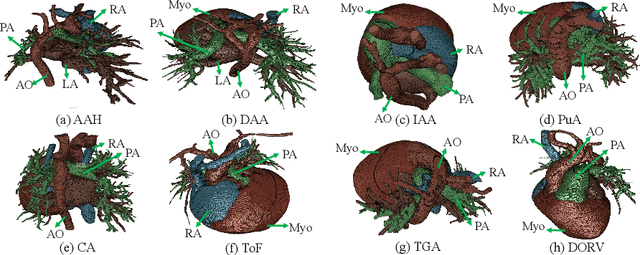
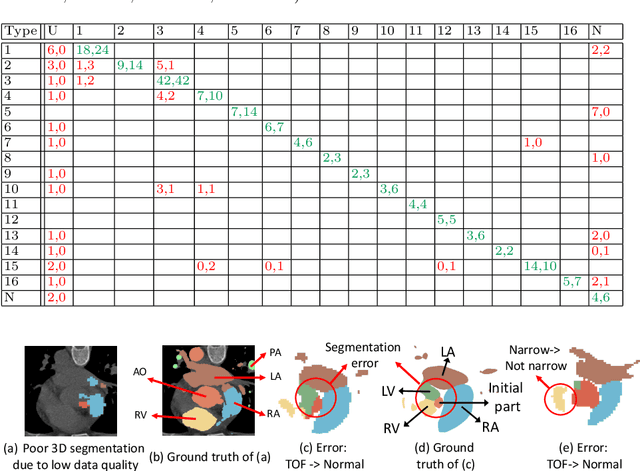
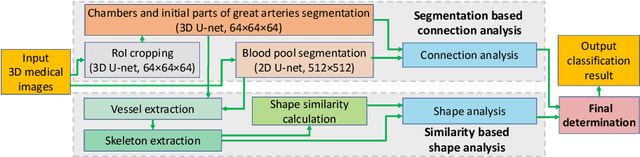
Abstract:Congenital heart disease (CHD) is the most common type of birth defect, which occurs 1 in every 110 births in the United States. CHD usually comes with severe variations in heart structure and great artery connections that can be classified into many types. Thus highly specialized domain knowledge and the time-consuming human process is needed to analyze the associated medical images. On the other hand, due to the complexity of CHD and the lack of dataset, little has been explored on the automatic diagnosis (classification) of CHDs. In this paper, we present ImageCHD, the first medical image dataset for CHD classification. ImageCHD contains 110 3D Computed Tomography (CT) images covering most types of CHD, which is of decent size Classification of CHDs requires the identification of large structural changes without any local tissue changes, with limited data. It is an example of a larger class of problems that are quite difficult for current machine-learning-based vision methods to solve. To demonstrate this, we further present a baseline framework for the automatic classification of CHD, based on a state-of-the-art CHD segmentation method. Experimental results show that the baseline framework can only achieve a classification accuracy of 82.0\% under a selective prediction scheme with 88.4\% coverage, leaving big room for further improvement. We hope that ImageCHD can stimulate further research and lead to innovative and generic solutions that would have an impact in multiple domains. Our dataset is released to the public compared with existing medical imaging datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge