Xiaobo Zhang
School of Information Engineering, Jiangxi Vocational College of Finance & Economics, Jiujiang, China, School of Computer Sciences, Universiti Sains Malaysia, Penang, Malaysia
ESNERA: Empirical and semantic named entity alignment for named entity dataset merging
Aug 09, 2025Abstract:Named Entity Recognition (NER) is a fundamental task in natural language processing. It remains a research hotspot due to its wide applicability across domains. Although recent advances in deep learning have significantly improved NER performance, they rely heavily on large, high-quality annotated datasets. However, building these datasets is expensive and time-consuming, posing a major bottleneck for further research. Current dataset merging approaches mainly focus on strategies like manual label mapping or constructing label graphs, which lack interpretability and scalability. To address this, we propose an automatic label alignment method based on label similarity. The method combines empirical and semantic similarities, using a greedy pairwise merging strategy to unify label spaces across different datasets. Experiments are conducted in two stages: first, merging three existing NER datasets into a unified corpus with minimal impact on NER performance; second, integrating this corpus with a small-scale, self-built dataset in the financial domain. The results show that our method enables effective dataset merging and enhances NER performance in the low-resource financial domain. This study presents an efficient, interpretable, and scalable solution for integrating multi-source NER corpora.
Dual-branch Prompting for Multimodal Machine Translation
Jul 23, 2025Abstract:Multimodal Machine Translation (MMT) typically enhances text-only translation by incorporating aligned visual features. Despite the remarkable progress, state-of-the-art MMT approaches often rely on paired image-text inputs at inference and are sensitive to irrelevant visual noise, which limits their robustness and practical applicability. To address these issues, we propose D2P-MMT, a diffusion-based dual-branch prompting framework for robust vision-guided translation. Specifically, D2P-MMT requires only the source text and a reconstructed image generated by a pre-trained diffusion model, which naturally filters out distracting visual details while preserving semantic cues. During training, the model jointly learns from both authentic and reconstructed images using a dual-branch prompting strategy, encouraging rich cross-modal interactions. To bridge the modality gap and mitigate training-inference discrepancies, we introduce a distributional alignment loss that enforces consistency between the output distributions of the two branches. Extensive experiments on the Multi30K dataset demonstrate that D2P-MMT achieves superior translation performance compared to existing state-of-the-art approaches.
Redefining Neural Operators in $d+1$ Dimensions
May 17, 2025Abstract:Neural Operators have emerged as powerful tools for learning mappings between function spaces. Among them, the kernel integral operator has been widely validated on universally approximating various operators. Although recent advancements following this definition have developed effective modules to better approximate the kernel function defined on the original domain (with $d$ dimensions, $d=1, 2, 3...$), the unclarified evolving mechanism in the embedding spaces blocks our view to design neural operators that can fully capture the target system evolution. Drawing on recent breakthroughs in quantum simulation of partial differential equations (PDEs), we elucidate the linear evolution process in neural operators. Based on that, we redefine neural operators on a new $d+1$ dimensional domain. Within this framework, we implement our proposed Schr\"odingerised Kernel Neural Operator (SKNO) aligning better with the $d+1$ dimensional evolution. In experiments, our $d+1$ dimensional evolving linear block performs far better than others. Also, we test SKNO's SOTA performance on various benchmark tests and also the zero-shot super-resolution task. In addition, we analyse the impact of different lifting and recovering operators on the prediction within the redefined NO framework, reflecting the alignment between our model and the underlying $d+1$ dimensional evolution.
AOR: Anatomical Ontology-Guided Reasoning for Medical Large Multimodal Model in Chest X-Ray Interpretation
May 05, 2025Abstract:Chest X-rays (CXRs) are the most frequently performed imaging examinations in clinical settings. Recent advancements in Large Multimodal Models (LMMs) have enabled automated CXR interpretation, enhancing diagnostic accuracy and efficiency. However, despite their strong visual understanding, current Medical LMMs (MLMMs) still face two major challenges: (1) Insufficient region-level understanding and interaction, and (2) Limited accuracy and interpretability due to single-step reasoning. In this paper, we empower MLMMs with anatomy-centric reasoning capabilities to enhance their interactivity and explainability. Specifically, we first propose an Anatomical Ontology-Guided Reasoning (AOR) framework, which centers on cross-modal region-level information to facilitate multi-step reasoning. Next, under the guidance of expert physicians, we develop AOR-Instruction, a large instruction dataset for MLMMs training. Our experiments demonstrate AOR's superior performance in both VQA and report generation tasks.
CT2C-QA: Multimodal Question Answering over Chinese Text, Table and Chart
Oct 28, 2024Abstract:Multimodal Question Answering (MMQA) is crucial as it enables comprehensive understanding and accurate responses by integrating insights from diverse data representations such as tables, charts, and text. Most existing researches in MMQA only focus on two modalities such as image-text QA, table-text QA and chart-text QA, and there remains a notable scarcity in studies that investigate the joint analysis of text, tables, and charts. In this paper, we present C$\text{T}^2$C-QA, a pioneering Chinese reasoning-based QA dataset that includes an extensive collection of text, tables, and charts, meticulously compiled from 200 selectively sourced webpages. Our dataset simulates real webpages and serves as a great test for the capability of the model to analyze and reason with multimodal data, because the answer to a question could appear in various modalities, or even potentially not exist at all. Additionally, we present AED (\textbf{A}llocating, \textbf{E}xpert and \textbf{D}esicion), a multi-agent system implemented through collaborative deployment, information interaction, and collective decision-making among different agents. Specifically, the Assignment Agent is in charge of selecting and activating expert agents, including those proficient in text, tables, and charts. The Decision Agent bears the responsibility of delivering the final verdict, drawing upon the analytical insights provided by these expert agents. We execute a comprehensive analysis, comparing AED with various state-of-the-art models in MMQA, including GPT-4. The experimental outcomes demonstrate that current methodologies, including GPT-4, are yet to meet the benchmarks set by our dataset.
A Medical Multimodal Large Language Model for Pediatric Pneumonia
Sep 04, 2024



Abstract:Pediatric pneumonia is the leading cause of death among children under five years worldwide, imposing a substantial burden on affected families. Currently, there are three significant hurdles in diagnosing and treating pediatric pneumonia. Firstly, pediatric pneumonia shares similar symptoms with other respiratory diseases, making rapid and accurate differential diagnosis challenging. Secondly, primary hospitals often lack sufficient medical resources and experienced doctors. Lastly, providing personalized diagnostic reports and treatment recommendations is labor-intensive and time-consuming. To tackle these challenges, we proposed a Medical Multimodal Large Language Model for Pediatric Pneumonia (P2Med-MLLM). It was capable of handling diverse clinical tasks, such as generating free-text radiology reports and medical records within a unified framework. Specifically, P2Med-MLLM can process both pure text and image-text data, trained on an extensive and large-scale dataset (P2Med-MD), including real clinical information from 163,999 outpatient and 8,684 inpatient cases. This dataset comprised 2D chest X-ray images, 3D chest CT images, corresponding radiology reports, and outpatient and inpatient records. We designed a three-stage training strategy to enable P2Med-MLLM to comprehend medical knowledge and follow instructions for various clinical tasks. To rigorously evaluate P2Med-MLLM's performance, we developed P2Med-MBench, a benchmark consisting of 642 meticulously verified samples by pediatric pulmonology specialists, covering six clinical decision-support tasks and a balanced variety of diseases. The automated scoring results demonstrated the superiority of P2Med-MLLM. This work plays a crucial role in assisting primary care doctors with prompt disease diagnosis and treatment planning, reducing severe symptom mortality rates, and optimizing the allocation of medical resources.
Anatomical Structure-Guided Medical Vision-Language Pre-training
Mar 14, 2024Abstract:Learning medical visual representations through vision-language pre-training has reached remarkable progress. Despite the promising performance, it still faces challenges, i.e., local alignment lacks interpretability and clinical relevance, and the insufficient internal and external representation learning of image-report pairs. To address these issues, we propose an Anatomical Structure-Guided (ASG) framework. Specifically, we parse raw reports into triplets <anatomical region, finding, existence>, and fully utilize each element as supervision to enhance representation learning. For anatomical region, we design an automatic anatomical region-sentence alignment paradigm in collaboration with radiologists, considering them as the minimum semantic units to explore fine-grained local alignment. For finding and existence, we regard them as image tags, applying an image-tag recognition decoder to associate image features with their respective tags within each sample and constructing soft labels for contrastive learning to improve the semantic association of different image-report pairs. We evaluate the proposed ASG framework on two downstream tasks, including five public benchmarks. Experimental results demonstrate that our method outperforms the state-of-the-art methods.
Let All be Whitened: Multi-teacher Distillation for Efficient Visual Retrieval
Dec 15, 2023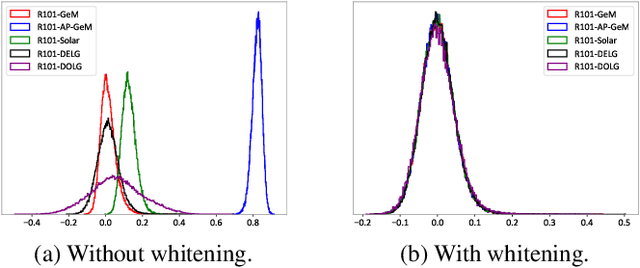
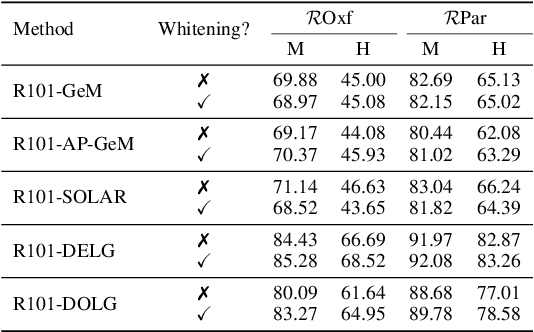
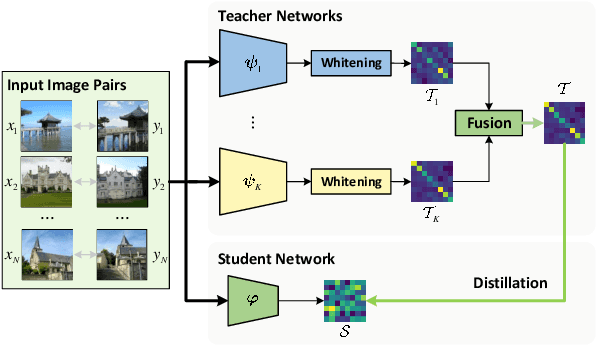
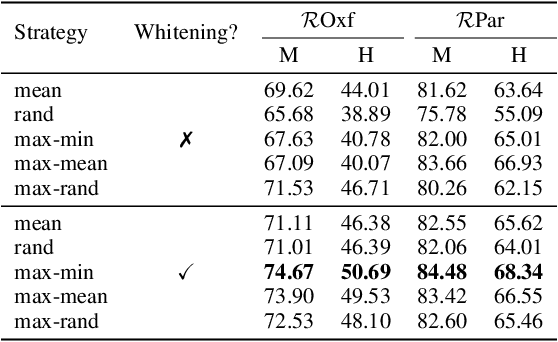
Abstract:Visual retrieval aims to search for the most relevant visual items, e.g., images and videos, from a candidate gallery with a given query item. Accuracy and efficiency are two competing objectives in retrieval tasks. Instead of crafting a new method pursuing further improvement on accuracy, in this paper we propose a multi-teacher distillation framework Whiten-MTD, which is able to transfer knowledge from off-the-shelf pre-trained retrieval models to a lightweight student model for efficient visual retrieval. Furthermore, we discover that the similarities obtained by different retrieval models are diversified and incommensurable, which makes it challenging to jointly distill knowledge from multiple models. Therefore, we propose to whiten the output of teacher models before fusion, which enables effective multi-teacher distillation for retrieval models. Whiten-MTD is conceptually simple and practically effective. Extensive experiments on two landmark image retrieval datasets and one video retrieval dataset demonstrate the effectiveness of our proposed method, and its good balance of retrieval performance and efficiency. Our source code is released at https://github.com/Maryeon/whiten_mtd.
Large Language Models are Complex Table Parsers
Dec 13, 2023



Abstract:With the Generative Pre-trained Transformer 3.5 (GPT-3.5) exhibiting remarkable reasoning and comprehension abilities in Natural Language Processing (NLP), most Question Answering (QA) research has primarily centered around general QA tasks based on GPT, neglecting the specific challenges posed by Complex Table QA. In this paper, we propose to incorporate GPT-3.5 to address such challenges, in which complex tables are reconstructed into tuples and specific prompt designs are employed for dialogues. Specifically, we encode each cell's hierarchical structure, position information, and content as a tuple. By enhancing the prompt template with an explanatory description of the meaning of each tuple and the logical reasoning process of the task, we effectively improve the hierarchical structure awareness capability of GPT-3.5 to better parse the complex tables. Extensive experiments and results on Complex Table QA datasets, i.e., the open-domain dataset HiTAB and the aviation domain dataset AIT-QA show that our approach significantly outperforms previous work on both datasets, leading to state-of-the-art (SOTA) performance.
Enhanced Knowledge Injection for Radiology Report Generation
Nov 01, 2023



Abstract:Automatic generation of radiology reports holds crucial clinical value, as it can alleviate substantial workload on radiologists and remind less experienced ones of potential anomalies. Despite the remarkable performance of various image captioning methods in the natural image field, generating accurate reports for medical images still faces challenges, i.e., disparities in visual and textual data, and lack of accurate domain knowledge. To address these issues, we propose an enhanced knowledge injection framework, which utilizes two branches to extract different types of knowledge. The Weighted Concept Knowledge (WCK) branch is responsible for introducing clinical medical concepts weighted by TF-IDF scores. The Multimodal Retrieval Knowledge (MRK) branch extracts triplets from similar reports, emphasizing crucial clinical information related to entity positions and existence. By integrating this finer-grained and well-structured knowledge with the current image, we are able to leverage the multi-source knowledge gain to ultimately facilitate more accurate report generation. Extensive experiments have been conducted on two public benchmarks, demonstrating that our method achieves superior performance over other state-of-the-art methods. Ablation studies further validate the effectiveness of two extracted knowledge sources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge