Stefano B. Blumberg
Centre for Medical Image Computing and Department of Computer Science - University College London - UK
Low-field magnetic resonance image enhancement via stochastic image quality transfer
Apr 26, 2023



Abstract:Low-field (<1T) magnetic resonance imaging (MRI) scanners remain in widespread use in low- and middle-income countries (LMICs) and are commonly used for some applications in higher income countries e.g. for small child patients with obesity, claustrophobia, implants, or tattoos. However, low-field MR images commonly have lower resolution and poorer contrast than images from high field (1.5T, 3T, and above). Here, we present Image Quality Transfer (IQT) to enhance low-field structural MRI by estimating from a low-field image the image we would have obtained from the same subject at high field. Our approach uses (i) a stochastic low-field image simulator as the forward model to capture uncertainty and variation in the contrast of low-field images corresponding to a particular high-field image, and (ii) an anisotropic U-Net variant specifically designed for the IQT inverse problem. We evaluate the proposed algorithm both in simulation and using multi-contrast (T1-weighted, T2-weighted, and fluid attenuated inversion recovery (FLAIR)) clinical low-field MRI data from an LMIC hospital. We show the efficacy of IQT in improving contrast and resolution of low-field MR images. We demonstrate that IQT-enhanced images have potential for enhancing visualisation of anatomical structures and pathological lesions of clinical relevance from the perspective of radiologists. IQT is proved to have capability of boosting the diagnostic value of low-field MRI, especially in low-resource settings.
Deformably-Scaled Transposed Convolution
Oct 17, 2022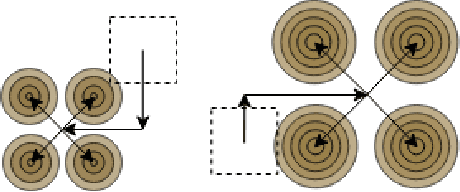

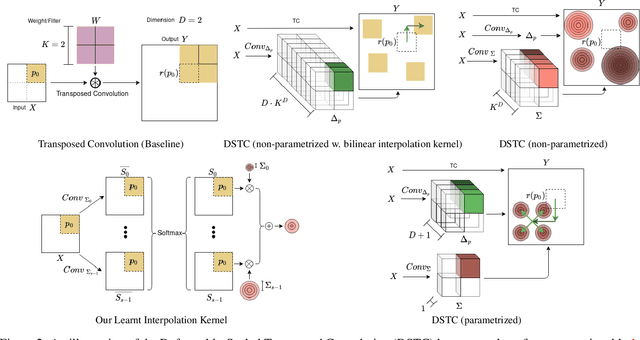
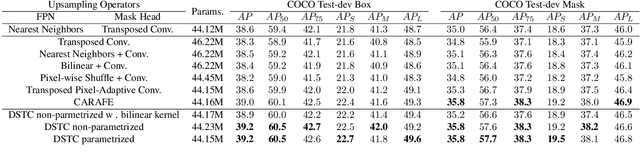
Abstract:Transposed convolution is crucial for generating high-resolution outputs, yet has received little attention compared to convolution layers. In this work we revisit transposed convolution and introduce a novel layer that allows us to place information in the image selectively and choose the `stroke breadth' at which the image is synthesized, whilst incurring a small additional parameter cost. For this we introduce three ideas: firstly, we regress offsets to the positions where the transpose convolution results are placed; secondly we broadcast the offset weight locations over a learnable neighborhood; and thirdly we use a compact parametrization to share weights and restrict offsets. We show that simply substituting upsampling operators with our novel layer produces substantial improvements across tasks as diverse as instance segmentation, object detection, semantic segmentation, generative image modeling, and 3D magnetic resonance image enhancement, while outperforming all existing variants of transposed convolutions. Our novel layer can be used as a drop-in replacement for 2D and 3D upsampling operators and the code will be publicly available.
An Experiment Design Paradigm using Joint Feature Selection and Task Optimization
Oct 13, 2022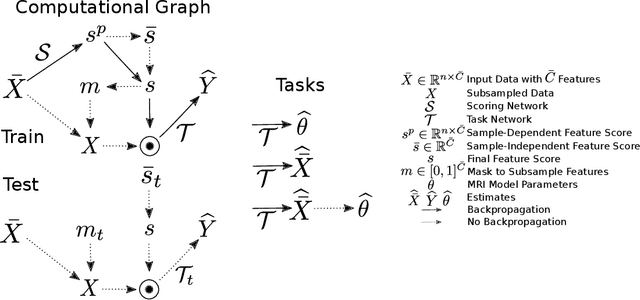
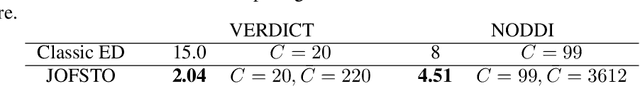
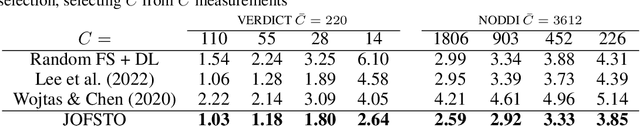
Abstract:This paper presents a subsampling-task paradigm for data-driven task-specific experiment design (ED) and a novel method in populationwide supervised feature selection (FS). Optimal ED, the choice of sampling points under constraints of limited acquisition-time, arises in a wide variety of scientific and engineering contexts. However the continuous optimization used in classical approaches depend on a-priori parameter choices and challenging non-convex optimization landscapes. This paper proposes to replace this strategy with a subsampling-task paradigm, analogous to populationwide supervised FS. In particular, we introduce JOFSTO, which performs JOint Feature Selection and Task Optimization. JOFSTO jointly optimizes two coupled networks: one for feature scoring, which provides the ED, the other for execution of a downstream task or process. Unlike most FS problems, e.g. selecting protein expressions for classification, ED problems typically select from highly correlated globally informative candidates rather than seeking a small number of highly informative features among many uninformative features. JOFSTO's construction efficiently identifies potentially correlated, but effective subsets and returns a trained task network. We demonstrate the approach using parameter estimation and mapping problems in quantitative MRI, where economical ED is crucial for clinical application. Results from simulations and empirical data show the subsampling-task paradigm strongly outperforms classical ED, and within our paradigm, JOFSTO outperforms state-of-the-art supervised FS techniques. JOFSTO extends immediately to wider image-based ED problems and other scenarios where the design must be specified globally across large numbers of acquisitions. Code will be released.
Fitting a Directional Microstructure Model to Diffusion-Relaxation MRI Data with Self-Supervised Machine Learning
Oct 05, 2022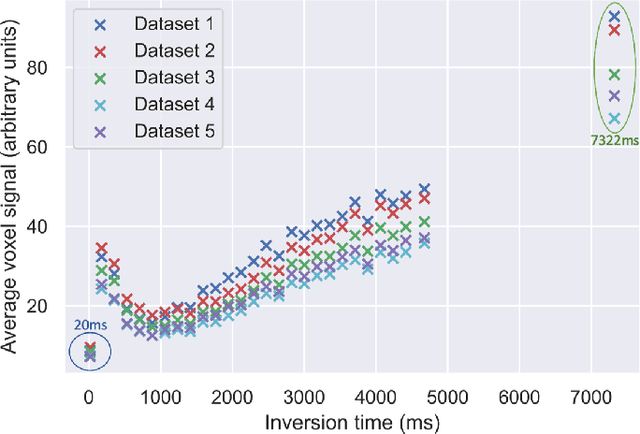
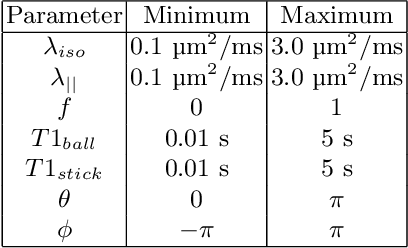
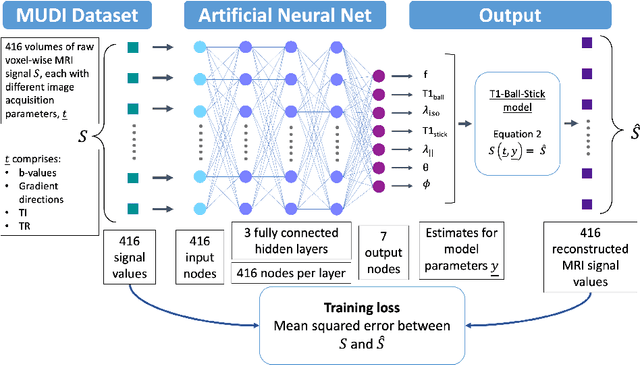
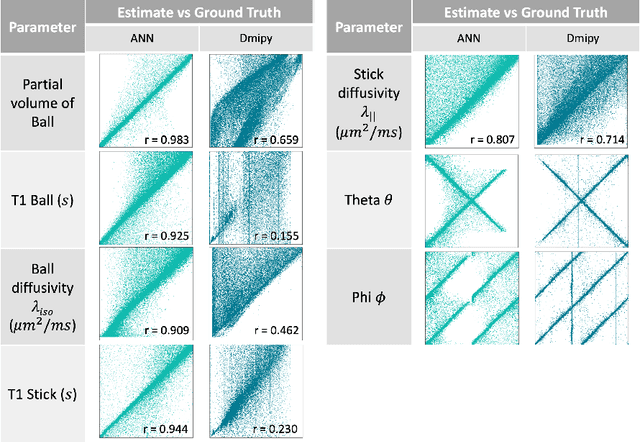
Abstract:Machine learning is a powerful approach for fitting microstructural models to diffusion MRI data. Early machine learning microstructure imaging implementations trained regressors to estimate model parameters in a supervised way, using synthetic training data with known ground truth. However, a drawback of this approach is that the choice of training data impacts fitted parameter values. Self-supervised learning is emerging as an attractive alternative to supervised learning in this context. Thus far, both supervised and self-supervised learning have typically been applied to isotropic models, such as intravoxel incoherent motion (IVIM), as opposed to models where the directionality of anisotropic structures is also estimated. In this paper, we demonstrate self-supervised machine learning model fitting for a directional microstructural model. In particular, we fit a combined T1-ball-stick model to the multidimensional diffusion (MUDI) challenge diffusion-relaxation dataset. Our self-supervised approach shows clear improvements in parameter estimation and computational time, for both simulated and in-vivo brain data, compared to standard non-linear least squares fitting. Code for the artificial neural net constructed for this study is available for public use from the following GitHub repository: https://github.com/jplte/deep-T1-ball-stick
Progressive Subsampling for Oversampled Data -- Application to Quantitative MRI
Apr 08, 2022
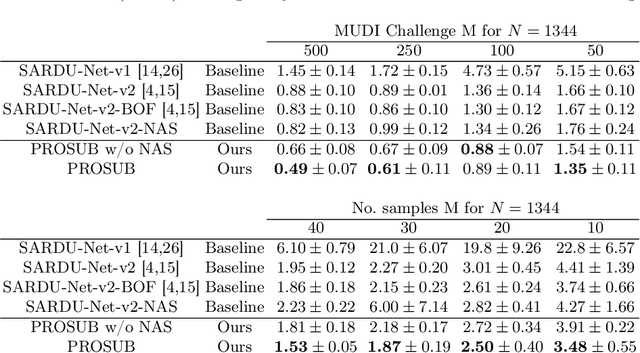
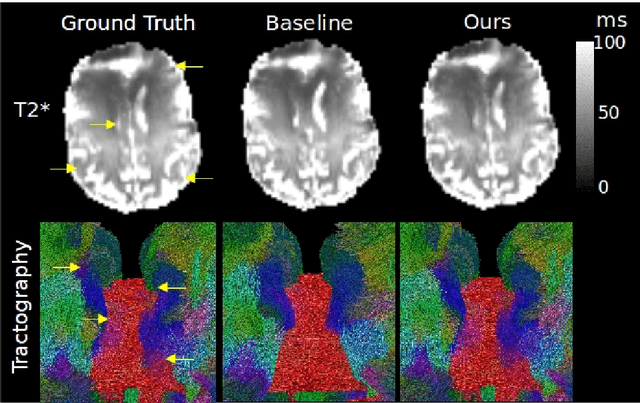
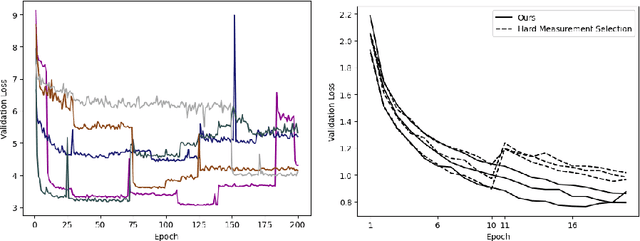
Abstract:We present PROSUB: PROgressive SUBsampling, a deep learning based, automated methodology that subsamples an oversampled data set (e.g. multi-channeled 3D images) with minimal loss of information. We build upon a recent dual-network approach that won the MICCAI MUlti-DIffusion (MUDI) quantitative MRI measurement sampling-reconstruction challenge, but suffers from deep learning training instability, by subsampling with a hard decision boundary. PROSUB uses the paradigm of recursive feature elimination (RFE) and progressively subsamples measurements during deep learning training, improving optimization stability. PROSUB also integrates a neural architecture search (NAS) paradigm, allowing the network architecture hyperparameters to respond to the subsampling process. We show PROSUB outperforms the winner of the MUDI MICCAI challenge, producing large improvements >18% MSE on the MUDI challenge sub-tasks and qualitative improvements on downstream processes useful for clinical applications. We also show the benefits of incorporating NAS and analyze the effect of PROSUB's components. As our method generalizes to other problems beyond MRI measurement selection-reconstruction, our code is https://github.com/sbb-gh/PROSUB
VAFO-Loss: VAscular Feature Optimised Loss Function for Retinal Artery/Vein Segmentation
Mar 12, 2022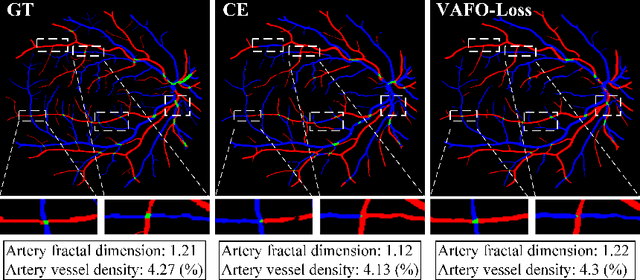
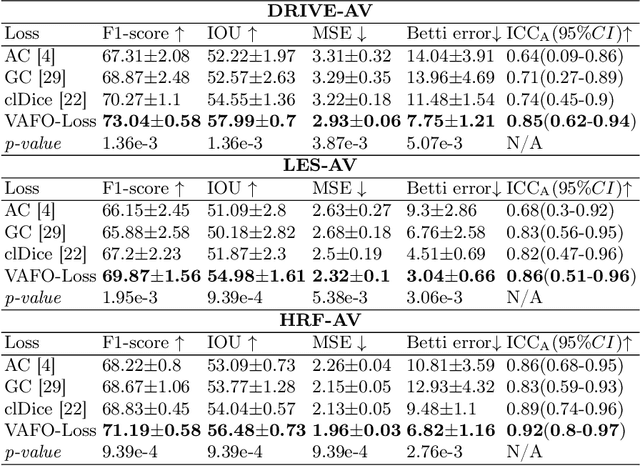
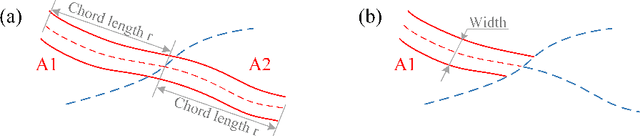
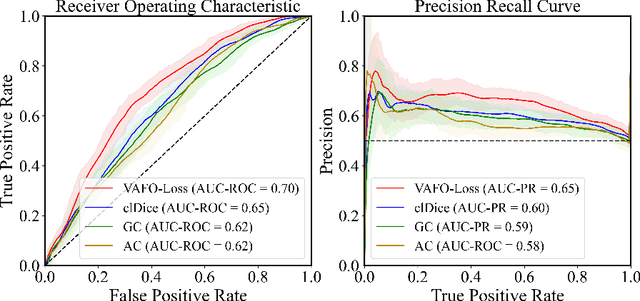
Abstract:Estimating clinically-relevant vascular features following vessel segmentation is a standard pipeline for retinal vessel analysis, which provides potential ocular biomarkers for both ophthalmic disease and systemic disease. In this work, we integrate these clinical features into a novel vascular feature optimised loss function (VAFO-Loss), in order to regularise networks to produce segmentation maps, with which more accurate vascular features can be derived. Two common vascular features, vessel density and fractal dimension, are identified to be sensitive to intra-segment misclassification, which is a well-recognised problem in multi-class artery/vein segmentation particularly hindering the estimation of these vascular features. Thus we encode these two features into VAFO-Loss. We first show that incorporating our end-to-end VAFO-Loss in standard segmentation networks indeed improves vascular feature estimation, yielding quantitative improvement in stroke incidence prediction, a clinical downstream task. We also report a technically interesting finding that the trained segmentation network, albeit biased by the feature optimised loss VAFO-Loss, shows statistically significant improvement in segmentation metrics, compared to those trained with other state-of-the-art segmentation losses.
MisMatch: Learning to Change Predictive Confidences with Attention for Consistency-Based, Semi-Supervised Medical Image Segmentation
Oct 23, 2021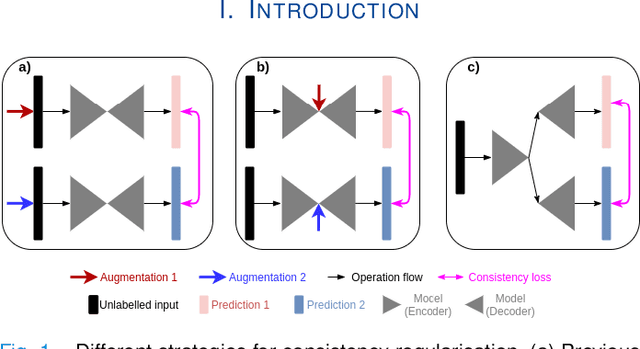
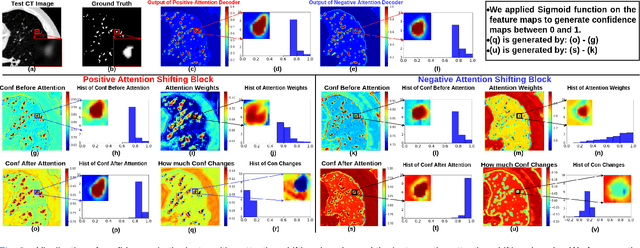

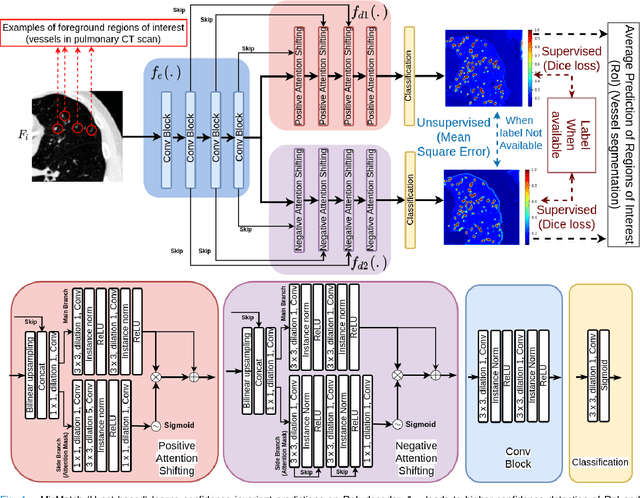
Abstract:The lack of labels is one of the fundamental constraints in deep learning based methods for image classification and segmentation, especially in applications such as medical imaging. Semi-supervised learning (SSL) is a promising method to address the challenge of labels carcity. The state-of-the-art SSL methods utilise consistency regularisation to learn unlabelled predictions which are invariant to perturbations on the prediction confidence. However, such SSL approaches rely on hand-crafted augmentation techniques which could be sub-optimal. In this paper, we propose MisMatch, a novel consistency based semi-supervised segmentation method. MisMatch automatically learns to produce paired predictions with increasedand decreased confidences. MisMatch consists of an encoder and two decoders. One decoder learns positive attention for regions of interest (RoI) on unlabelled data thereby generating higher confidence predictions of RoI. The other decoder learns negative attention for RoI on the same unlabelled data thereby generating lower confidence predictions. We then apply a consistency regularisation between the paired predictions of the decoders. For evaluation, we first perform extensive cross-validation on a CT-based pulmonary vessel segmentation task and show that MisMatch statistically outperforms state-of-the-art semi-supervised methods when only 6.25% of the total labels are used. Furthermore MisMatch performance using 6.25% ofthe total labels is comparable to state-of-the-art methodsthat utilise all available labels. In a second experiment, MisMatch outperforms state-of-the-art methods on an MRI-based brain tumour segmentation task.
DeepReg: a deep learning toolkit for medical image registration
Nov 04, 2020Abstract:DeepReg (https://github.com/DeepRegNet/DeepReg) is a community-supported open-source toolkit for research and education in medical image registration using deep learning.
QuantNet: Transferring Learning Across Systematic Trading Strategies
Apr 07, 2020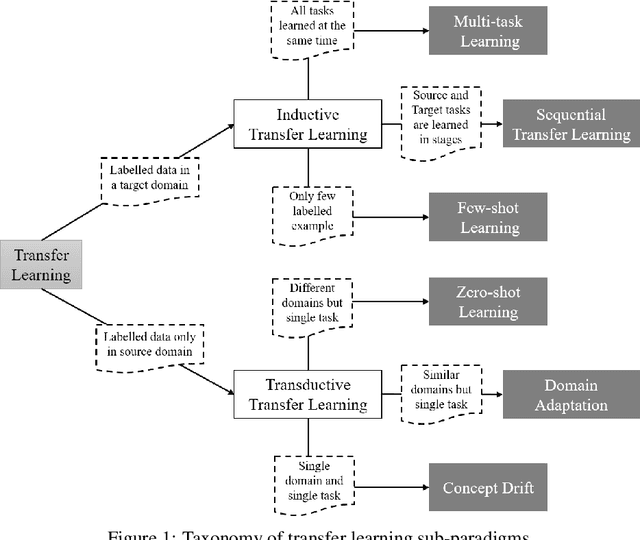
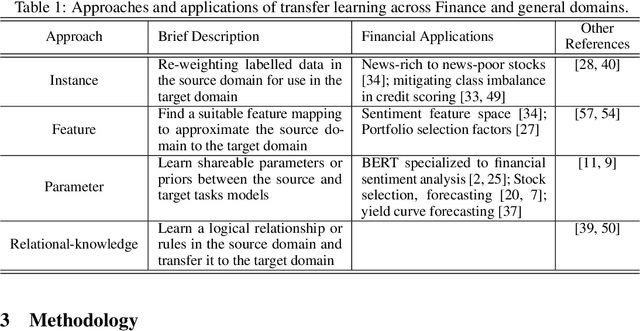
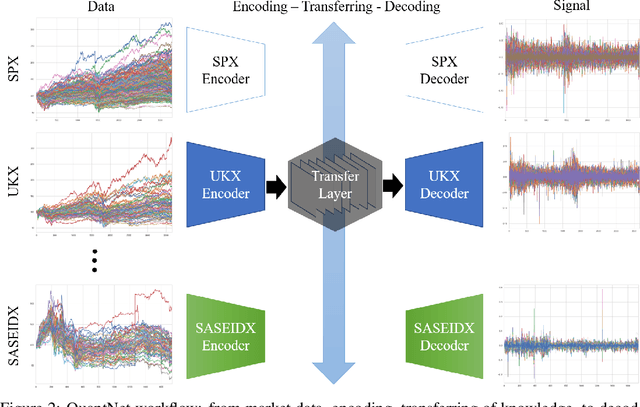
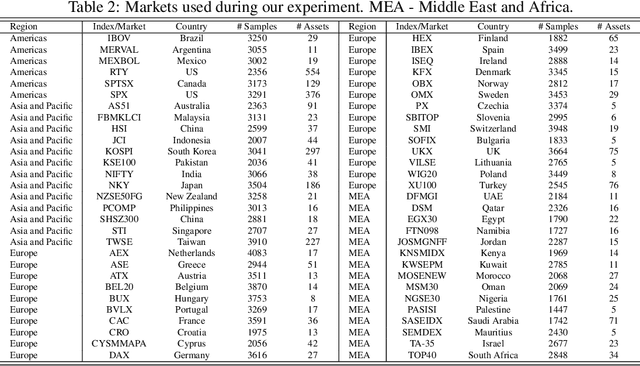
Abstract:In this work we introduce QuantNet: an architecture that is capable of transferring knowledge over systematic trading strategies in several financial markets. By having a system that is able to leverage and share knowledge across them, our aim is two-fold: to circumvent the so-called Backtest Overfitting problem; and to generate higher risk-adjusted returns and fewer drawdowns. To do that, QuantNet exploits a form of modelling called Transfer Learning, where two layers are market-specific and another one is market-agnostic. This ensures that the transfer occurs across trading strategies, with the market-agnostic layer acting as a vehicle to share knowledge, cross-influence each strategy parameters, and ultimately the trading signal produced. In order to evaluate QuantNet, we compared its performance in relation to the option of not performing transfer learning, that is, using market-specific old-fashioned machine learning. In summary, our findings suggest that QuantNet performs better than non transfer-based trading strategies, improving Sharpe ratio in 15% and Calmar ratio in 41% across 3103 assets in 58 equity markets across the world. Code coming soon.
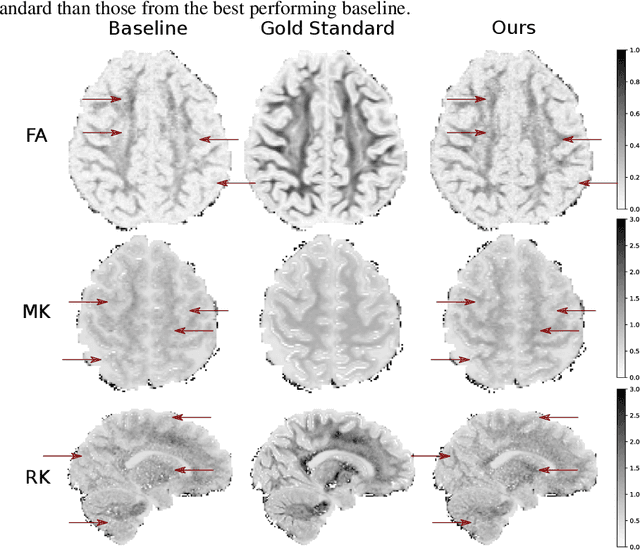
Image Quality Transfer Enhances Contrast and Resolution of Low-Field Brain MRI in African Paediatric Epilepsy Patients
Mar 18, 2020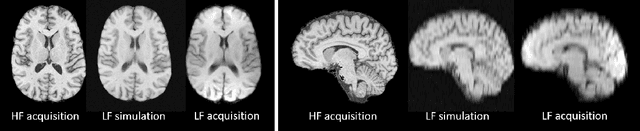
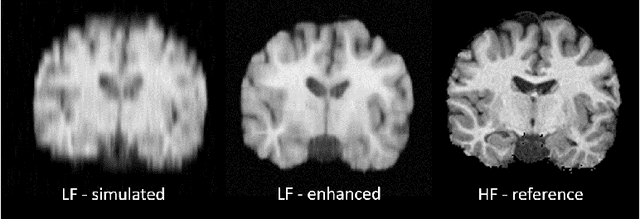
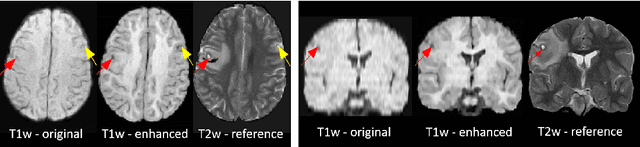
Abstract:1.5T or 3T scanners are the current standard for clinical MRI, but low-field (<1T) scanners are still common in many lower- and middle-income countries for reasons of cost and robustness to power failures. Compared to modern high-field scanners, low-field scanners provide images with lower signal-to-noise ratio at equivalent resolution, leaving practitioners to compensate by using large slice thickness and incomplete spatial coverage. Furthermore, the contrast between different types of brain tissue may be substantially reduced even at equal signal-to-noise ratio, which limits diagnostic value. Recently the paradigm of Image Quality Transfer has been applied to enhance 0.36T structural images aiming to approximate the resolution, spatial coverage, and contrast of typical 1.5T or 3T images. A variant of the neural network U-Net was trained using low-field images simulated from the publicly available 3T Human Connectome Project dataset. Here we present qualitative results from real and simulated clinical low-field brain images showing the potential value of IQT to enhance the clinical utility of readily accessible low-field MRIs in the management of epilepsy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge