Biobele J. Brown
Department of Paediatrics - College of Medicine - University of Ibadan - Nigeria
Low-field magnetic resonance image enhancement via stochastic image quality transfer
Apr 26, 2023



Abstract:Low-field (<1T) magnetic resonance imaging (MRI) scanners remain in widespread use in low- and middle-income countries (LMICs) and are commonly used for some applications in higher income countries e.g. for small child patients with obesity, claustrophobia, implants, or tattoos. However, low-field MR images commonly have lower resolution and poorer contrast than images from high field (1.5T, 3T, and above). Here, we present Image Quality Transfer (IQT) to enhance low-field structural MRI by estimating from a low-field image the image we would have obtained from the same subject at high field. Our approach uses (i) a stochastic low-field image simulator as the forward model to capture uncertainty and variation in the contrast of low-field images corresponding to a particular high-field image, and (ii) an anisotropic U-Net variant specifically designed for the IQT inverse problem. We evaluate the proposed algorithm both in simulation and using multi-contrast (T1-weighted, T2-weighted, and fluid attenuated inversion recovery (FLAIR)) clinical low-field MRI data from an LMIC hospital. We show the efficacy of IQT in improving contrast and resolution of low-field MR images. We demonstrate that IQT-enhanced images have potential for enhancing visualisation of anatomical structures and pathological lesions of clinical relevance from the perspective of radiologists. IQT is proved to have capability of boosting the diagnostic value of low-field MRI, especially in low-resource settings.
Automated Detection of Acute Promyelocytic Leukemia in Blood Films and Bone Marrow Aspirates with Annotation-free Deep Learning
Mar 20, 2022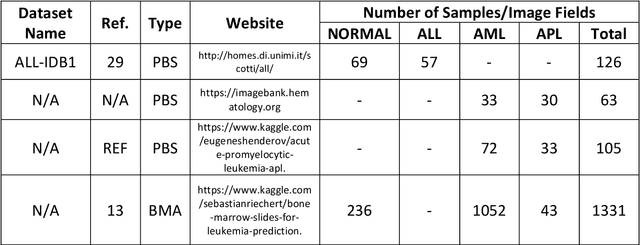
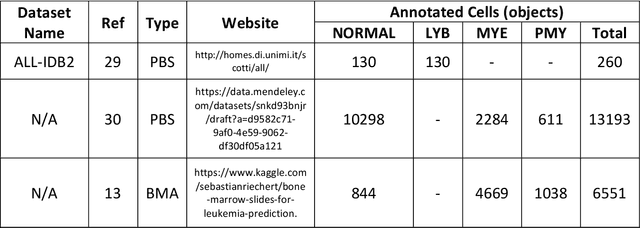
Abstract:While optical microscopy inspection of blood films and bone marrow aspirates by a hematologist is a crucial step in establishing diagnosis of acute leukemia, especially in low-resource settings where other diagnostic modalities might not be available, the task remains time-consuming and prone to human inconsistencies. This has an impact especially in cases of Acute Promyelocytic Leukemia (APL) that require urgent treatment. Integration of automated computational hematopathology into clinical workflows can improve the throughput of these services and reduce cognitive human error. However, a major bottleneck in deploying such systems is a lack of sufficient cell morphological object-labels annotations to train deep learning models. We overcome this by leveraging patient diagnostic labels to train weakly-supervised models that detect different types of acute leukemia. We introduce a deep learning approach, Multiple Instance Learning for Leukocyte Identification (MILLIE), able to perform automated reliable analysis of blood films with minimal supervision. Without being trained to classify individual cells, MILLIE differentiates between acute lymphoblastic and myeloblastic leukemia in blood films. More importantly, MILLIE detects APL in blood films (AUC 0.94+/-0.04) and in bone marrow aspirates (AUC 0.99+/-0.01). MILLIE is a viable solution to augment the throughput of clinical pathways that require assessment of blood film microscopy.
Image Quality Transfer Enhances Contrast and Resolution of Low-Field Brain MRI in African Paediatric Epilepsy Patients
Mar 18, 2020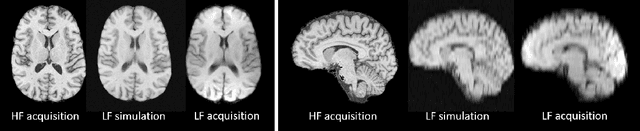
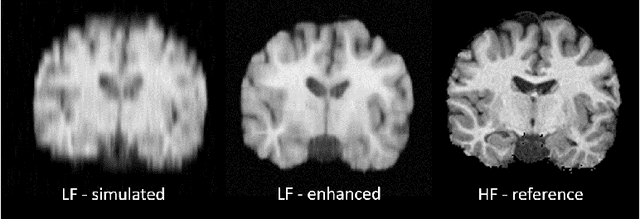
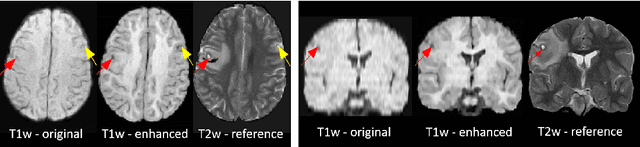
Abstract:1.5T or 3T scanners are the current standard for clinical MRI, but low-field (<1T) scanners are still common in many lower- and middle-income countries for reasons of cost and robustness to power failures. Compared to modern high-field scanners, low-field scanners provide images with lower signal-to-noise ratio at equivalent resolution, leaving practitioners to compensate by using large slice thickness and incomplete spatial coverage. Furthermore, the contrast between different types of brain tissue may be substantially reduced even at equal signal-to-noise ratio, which limits diagnostic value. Recently the paradigm of Image Quality Transfer has been applied to enhance 0.36T structural images aiming to approximate the resolution, spatial coverage, and contrast of typical 1.5T or 3T images. A variant of the neural network U-Net was trained using low-field images simulated from the publicly available 3T Human Connectome Project dataset. Here we present qualitative results from real and simulated clinical low-field brain images showing the potential value of IQT to enhance the clinical utility of readily accessible low-field MRIs in the management of epilepsy.
Deep Learning for Low-Field to High-Field MR: Image Quality Transfer with Probabilistic Decimation Simulator
Sep 15, 2019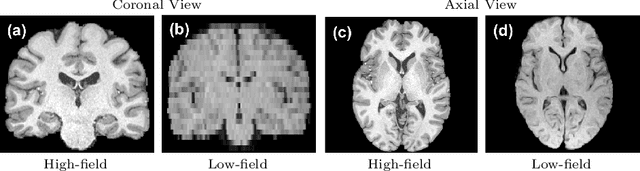
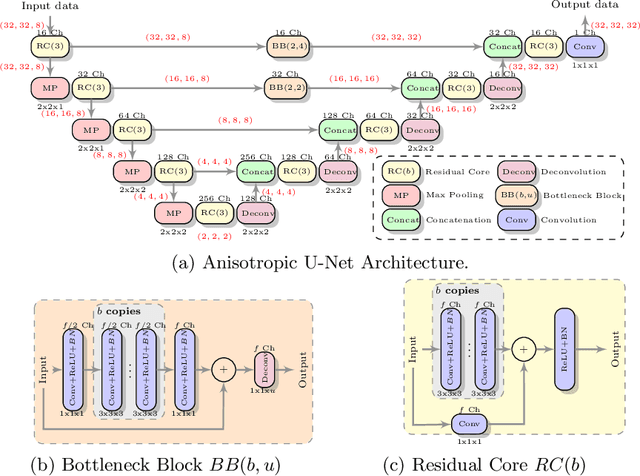
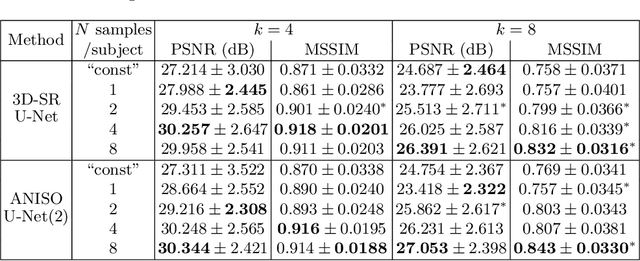
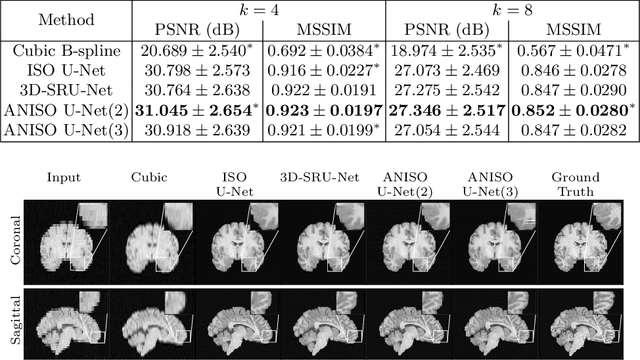
Abstract:MR images scanned at low magnetic field ($<1$T) have lower resolution in the slice direction and lower contrast, due to a relatively small signal-to-noise ratio (SNR) than those from high field (typically 1.5T and 3T). We adapt the recent idea of Image Quality Transfer (IQT) to enhance very low-field structural images aiming to estimate the resolution, spatial coverage, and contrast of high-field images. Analogous to many learning-based image enhancement techniques, IQT generates training data from high-field scans alone by simulating low-field images through a pre-defined decimation model. However, the ground truth decimation model is not well-known in practice, and lack of its specification can bias the trained model, aggravating performance on the real low-field scans. In this paper we propose a probabilistic decimation simulator to improve robustness of model training. It is used to generate and augment various low-field images whose parameters are random variables and sampled from an empirical distribution related to tissue-specific SNR on a 0.36T scanner. The probabilistic decimation simulator is model-agnostic, that is, it can be used with any super-resolution networks. Furthermore we propose a variant of U-Net architecture to improve its learning performance. We show promising qualitative results from clinical low-field images confirming the strong efficacy of IQT in an important new application area: epilepsy diagnosis in sub-Saharan Africa where only low-field scanners are normally available.
Data-Driven Malaria Prevalence Prediction in Large Densely-Populated Urban Holoendemic sub-Saharan West Africa: Harnessing Machine Learning Approaches and 22-years of Prospectively Collected Data
Jun 18, 2019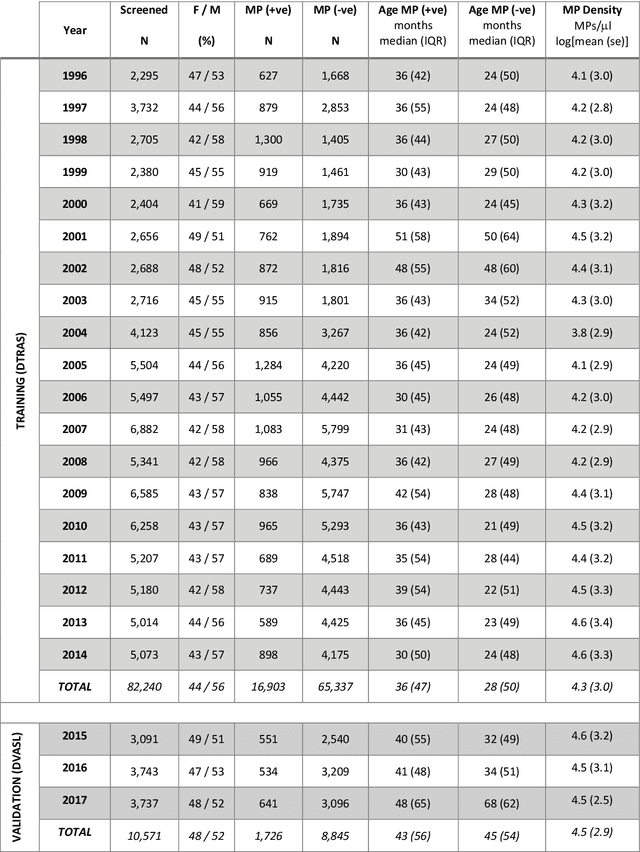
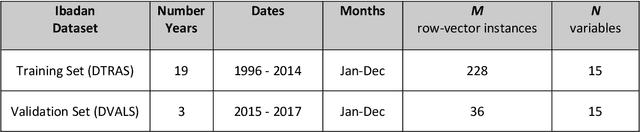
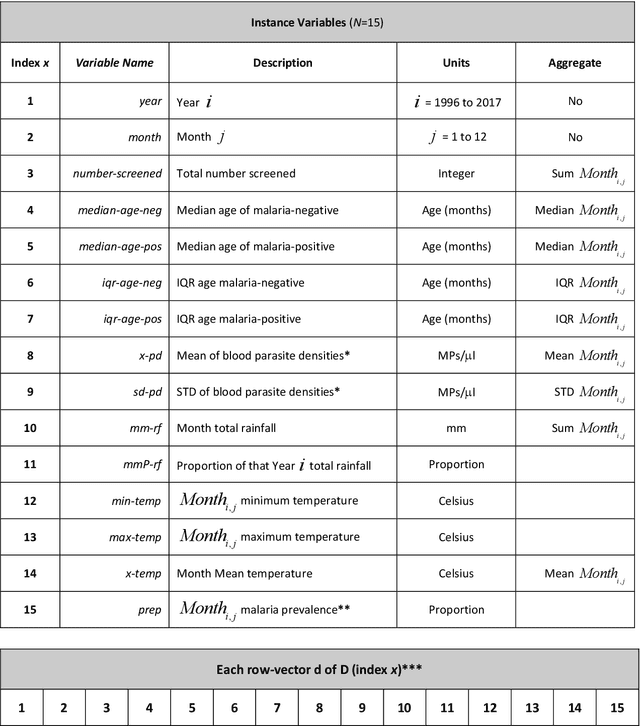
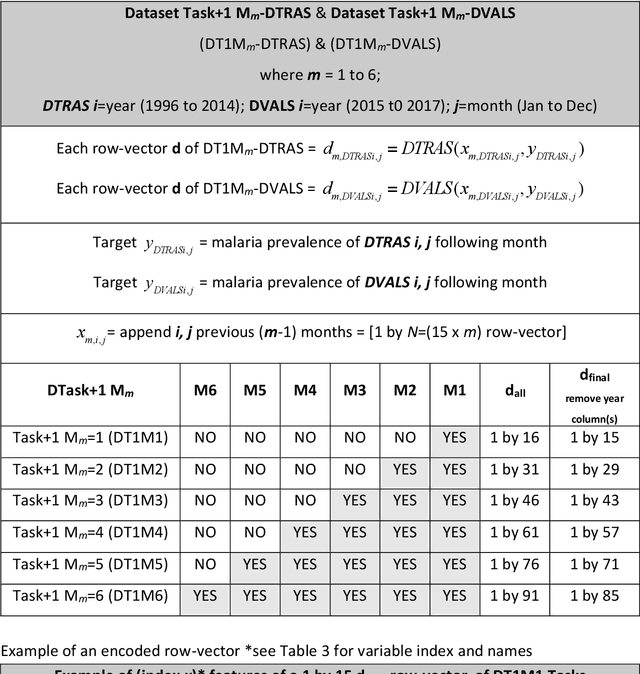
Abstract:Plasmodium falciparum malaria still poses one of the greatest threats to human life with over 200 million cases globally leading to half-million deaths annually. Of these, 90% of cases and of the mortality occurs in sub-Saharan Africa, mostly among children. Although malaria prediction systems are central to the 2016-2030 malaria Global Technical Strategy, currently these are inadequate at capturing and estimating the burden of disease in highly endemic countries. We developed and validated a computational system that exploits the predictive power of current Machine Learning approaches on 22-years of prospective data from the high-transmission holoendemic malaria urban-densely-populated sub-Saharan West-Africa metropolis of Ibadan. Our dataset of >9x104 screened study participants attending our clinical and community services from 1996 to 2017 contains monthly prevalence, temporal, environmental and host features. Our Locality-specific Elastic-Net based Malaria Prediction System (LEMPS) achieves good generalization performance, both in magnitude and direction of the prediction, when tasked to predict monthly prevalence on previously unseen validation data (MAE<=6x10-2, MSE<=7x10-3) within a range of (+0.1 to -0.05) error-tolerance which is relevant and usable for aiding decision-support in a holoendemic setting. LEMPS is well-suited for malaria prediction, where there are multiple features which are correlated with one another, and trading-off between regularization-strength L1-norm and L2-norm allows the system to retain stability. Data-driven systems are critical for regionally-adaptable surveillance, management of control strategies and resource allocation across stretched healthcare systems.
Deep Learning Enhanced Extended Depth-of-Field for Thick Blood-Film Malaria High-Throughput Microscopy
Jun 18, 2019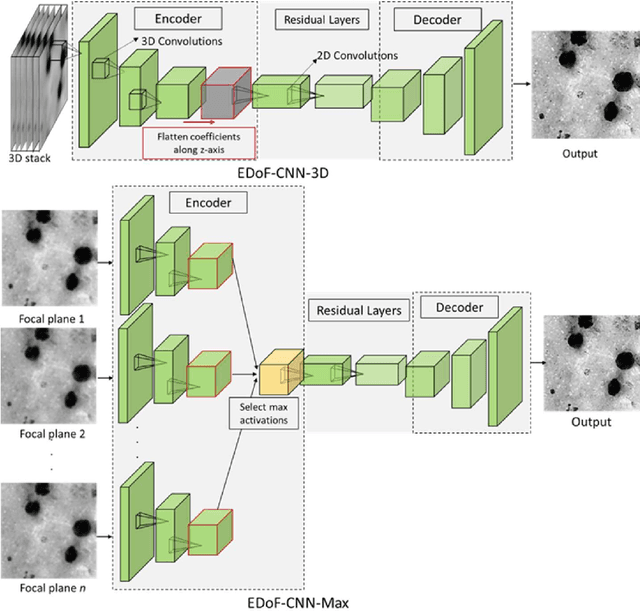
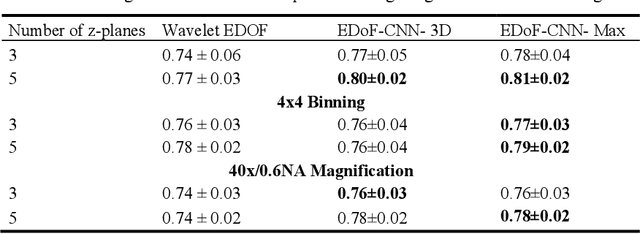
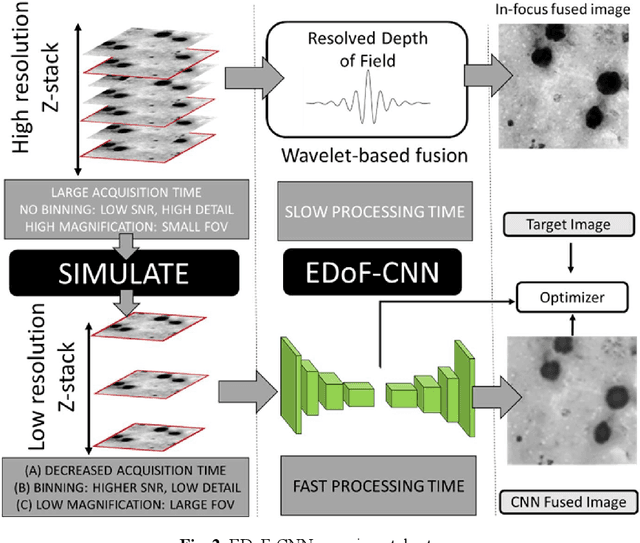
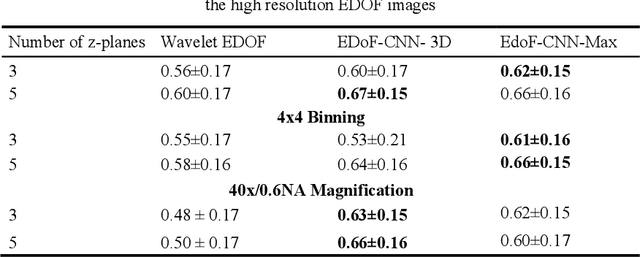
Abstract:Fast accurate diagnosis of malaria is still a global health challenge for which automated digital-pathology approaches could provide scalable solutions amenable to be deployed in low-to-middle income countries. Here we address the problem of Extended Depth-of-Field (EDoF) in thick blood film microscopy for rapid automated malaria diagnosis. High magnification oil-objectives (100x) with large numerical aperture are usually preferred to resolve the fine structural details that help separate true parasites from distractors. However, such objectives have a very limited depth-of-field requiring the acquisition of a series of images at different focal planes per field of view (FOV). Current EDoF techniques based on multi-scale decompositions are time consuming and therefore not suited for high-throughput analysis of specimens. To overcome this challenge, we developed a new deep learning method based on Convolutional Neural Networks (EDoF-CNN) that is able to rapidly perform the extended depth-of-field while also enhancing the spatial resolution of the resulting fused image. We evaluated our approach using simulated low-resolution z-stacks from Giemsa-stained thick blood smears from patients presenting with Plasmodium falciparum malaria. The EDoF-CNN allows speed-up of our digital-pathology acquisition platform and significantly improves the quality of the EDoF compared to the traditional multi-scaled approaches when applied to lower resolution stacks corresponding to acquisitions with fewer focal planes, large camera pixel binning or lower magnification objectives (larger FOV). We use the parasite detection accuracy of a deep learning model on the EDoFs as a concrete, task-specific measure of performance of this approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge