Enrico Kaden
Centre for Medical Image Computing and Department of Computer Science - University College London - UK, UCL Great Ormond Street Institute of Child Health - London - UK
Image Quality Transfer Enhances Contrast and Resolution of Low-Field Brain MRI in African Paediatric Epilepsy Patients
Mar 18, 2020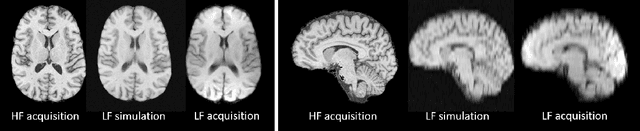
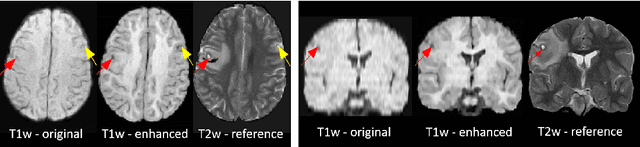
Abstract:1.5T or 3T scanners are the current standard for clinical MRI, but low-field (<1T) scanners are still common in many lower- and middle-income countries for reasons of cost and robustness to power failures. Compared to modern high-field scanners, low-field scanners provide images with lower signal-to-noise ratio at equivalent resolution, leaving practitioners to compensate by using large slice thickness and incomplete spatial coverage. Furthermore, the contrast between different types of brain tissue may be substantially reduced even at equal signal-to-noise ratio, which limits diagnostic value. Recently the paradigm of Image Quality Transfer has been applied to enhance 0.36T structural images aiming to approximate the resolution, spatial coverage, and contrast of typical 1.5T or 3T images. A variant of the neural network U-Net was trained using low-field images simulated from the publicly available 3T Human Connectome Project dataset. Here we present qualitative results from real and simulated clinical low-field brain images showing the potential value of IQT to enhance the clinical utility of readily accessible low-field MRIs in the management of epilepsy.
Deep Learning for Low-Field to High-Field MR: Image Quality Transfer with Probabilistic Decimation Simulator
Sep 15, 2019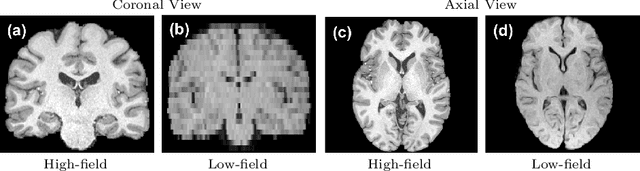
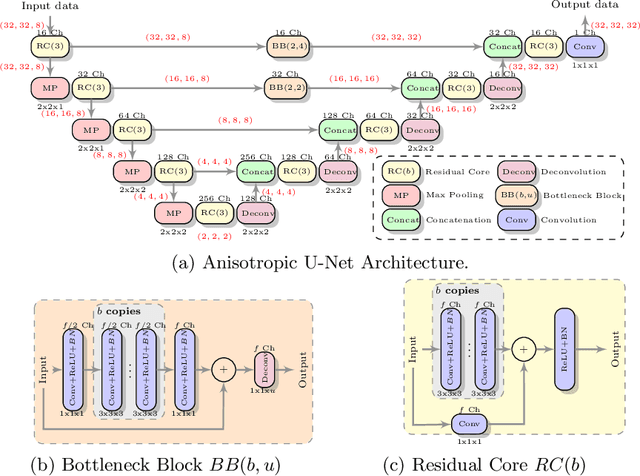
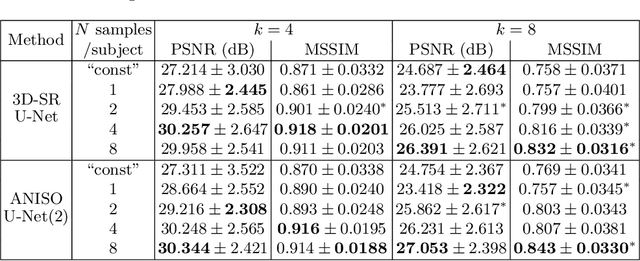
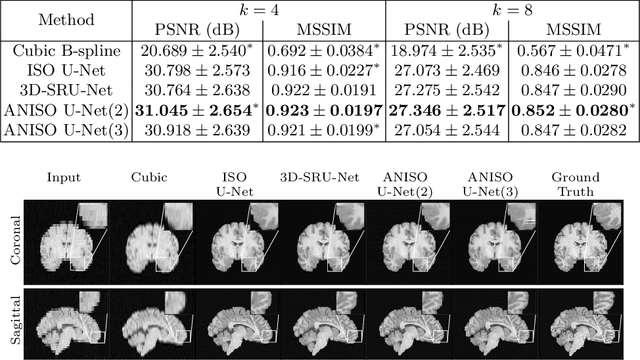
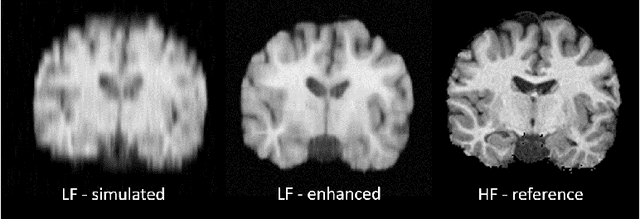
Abstract:MR images scanned at low magnetic field ($<1$T) have lower resolution in the slice direction and lower contrast, due to a relatively small signal-to-noise ratio (SNR) than those from high field (typically 1.5T and 3T). We adapt the recent idea of Image Quality Transfer (IQT) to enhance very low-field structural images aiming to estimate the resolution, spatial coverage, and contrast of high-field images. Analogous to many learning-based image enhancement techniques, IQT generates training data from high-field scans alone by simulating low-field images through a pre-defined decimation model. However, the ground truth decimation model is not well-known in practice, and lack of its specification can bias the trained model, aggravating performance on the real low-field scans. In this paper we propose a probabilistic decimation simulator to improve robustness of model training. It is used to generate and augment various low-field images whose parameters are random variables and sampled from an empirical distribution related to tissue-specific SNR on a 0.36T scanner. The probabilistic decimation simulator is model-agnostic, that is, it can be used with any super-resolution networks. Furthermore we propose a variant of U-Net architecture to improve its learning performance. We show promising qualitative results from clinical low-field images confirming the strong efficacy of IQT in an important new application area: epilepsy diagnosis in sub-Saharan Africa where only low-field scanners are normally available.
Uncertainty Quantification in Deep Learning for Safer Neuroimage Enhancement
Jul 31, 2019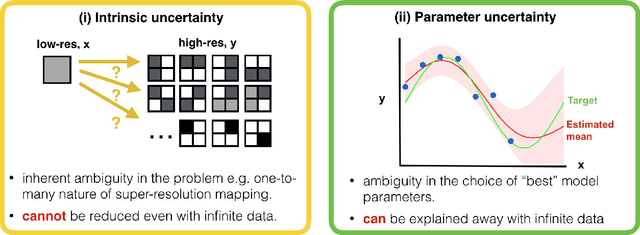
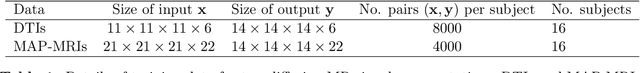
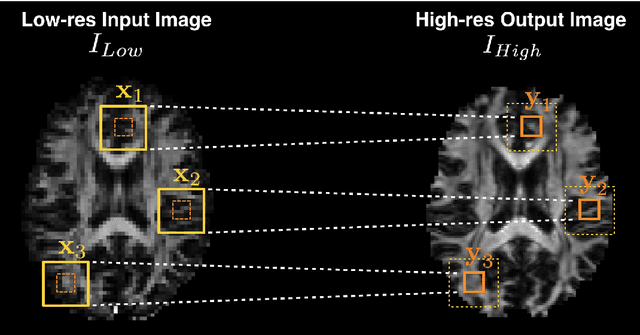
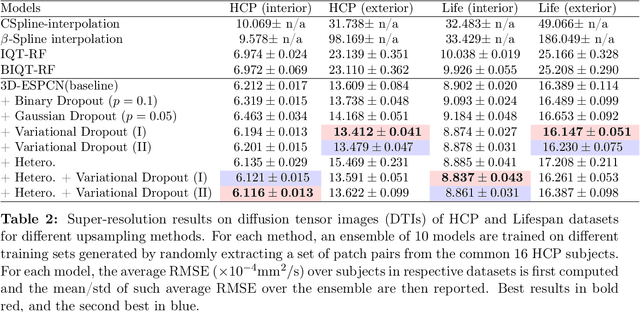
Abstract:Deep learning (DL) has shown great potential in medical image enhancement problems, such as super-resolution or image synthesis. However, to date, little consideration has been given to uncertainty quantification over the output image. Here we introduce methods to characterise different components of uncertainty in such problems and demonstrate the ideas using diffusion MRI super-resolution. Specifically, we propose to account for $intrinsic$ uncertainty through a heteroscedastic noise model and for $parameter$ uncertainty through approximate Bayesian inference, and integrate the two to quantify $predictive$ uncertainty over the output image. Moreover, we introduce a method to propagate the predictive uncertainty on a multi-channelled image to derived scalar parameters, and separately quantify the effects of intrinsic and parameter uncertainty therein. The methods are evaluated for super-resolution of two different signal representations of diffusion MR images---DTIs and Mean Apparent Propagator MRI---and their derived quantities such as MD and FA, on multiple datasets of both healthy and pathological human brains. Results highlight three key benefits of uncertainty modelling for improving the safety of DL-based image enhancement systems. Firstly, incorporating uncertainty improves the predictive performance even when test data departs from training data. Secondly, the predictive uncertainty highly correlates with errors, and is therefore capable of detecting predictive "failures". Results demonstrate that such an uncertainty measure enables subject-specific and voxel-wise risk assessment of the output images. Thirdly, we show that the method for decomposing predictive uncertainty into its independent sources provides high-level "explanations" for the performance by quantifying how much uncertainty arises from the inherent difficulty of the task or the limited training examples.
Bayesian Image Quality Transfer with CNNs: Exploring Uncertainty in dMRI Super-Resolution
May 30, 2017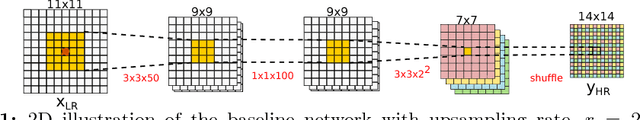
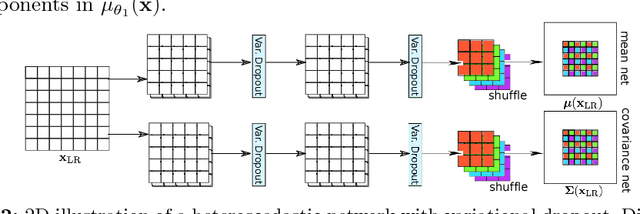
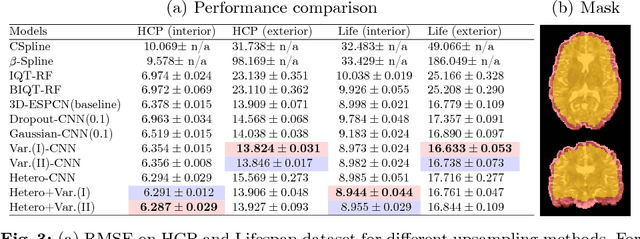
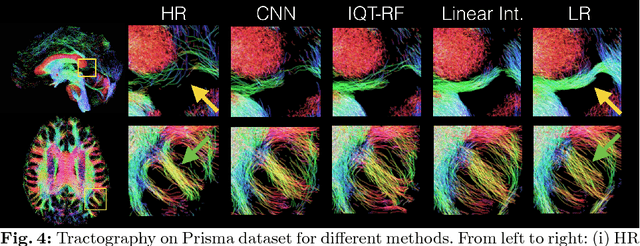
Abstract:In this work, we investigate the value of uncertainty modeling in 3D super-resolution with convolutional neural networks (CNNs). Deep learning has shown success in a plethora of medical image transformation problems, such as super-resolution (SR) and image synthesis. However, the highly ill-posed nature of such problems results in inevitable ambiguity in the learning of networks. We propose to account for intrinsic uncertainty through a per-patch heteroscedastic noise model and for parameter uncertainty through approximate Bayesian inference in the form of variational dropout. We show that the combined benefits of both lead to the state-of-the-art performance SR of diffusion MR brain images in terms of errors compared to ground truth. We further show that the reduced error scores produce tangible benefits in downstream tractography. In addition, the probabilistic nature of the methods naturally confers a mechanism to quantify uncertainty over the super-resolved output. We demonstrate through experiments on both healthy and pathological brains the potential utility of such an uncertainty measure in the risk assessment of the super-resolved images for subsequent clinical use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge