Ping Xu
Real-Time Lane Detection via Efficient Feature Alignment and Covariance Optimization for Low-Power Embedded Systems
Jan 05, 2026Abstract:Real-time lane detection in embedded systems encounters significant challenges due to subtle and sparse visual signals in RGB images, often constrained by limited computational resources and power consumption. Although deep learning models for lane detection categorized into segmentation-based, anchor-based, and curve-based methods there remains a scarcity of universally applicable optimization techniques tailored for low-power embedded environments. To overcome this, we propose an innovative Covariance Distribution Optimization (CDO) module specifically designed for efficient, real-time applications. The CDO module aligns lane feature distributions closely with ground-truth labels, significantly enhancing detection accuracy without increasing computational complexity. Evaluations were conducted on six diverse models across all three method categories, including two optimized for real-time applications and four state-of-the-art (SOTA) models, tested comprehensively on three major datasets: CULane, TuSimple, and LLAMAS. Experimental results demonstrate accuracy improvements ranging from 0.01% to 1.5%. The proposed CDO module is characterized by ease of integration into existing systems without structural modifications and utilizes existing model parameters to facilitate ongoing training, thus offering substantial benefits in performance, power efficiency, and operational flexibility in embedded systems.
DFedReweighting: A Unified Framework for Objective-Oriented Reweighting in Decentralized Federated Learning
Dec 12, 2025Abstract:Decentralized federated learning (DFL) has recently emerged as a promising paradigm that enables multiple clients to collaboratively train machine learning model through iterative rounds of local training, communication, and aggregation without relying on a central server which introduces potential vulnerabilities in conventional Federated Learning. Nevertheless, DFL systems continue to face a range of challenges, including fairness, robustness, etc. To address these challenges, we propose \textbf{DFedReweighting}, a unified aggregation framework designed to achieve diverse objectives in DFL systems via a objective-oriented reweighting aggregation at the final step of each learning round. Specifically, the framework first computes preliminary weights based on \textit{target performance metric} obtained from auxiliary dataset constructed using local data. These weights are then refined using \textit{customized reweighting strategy}, resulting in the final aggregation weights. Our results from the theoretical analysis demonstrate that the appropriate combination of the target performance metric and the customized reweighting strategy ensures linear convergence. Experimental results consistently show that our proposed framework significantly improves fairness and robustness against Byzantine attacks in diverse scenarios. Provided that appropriate target performance metrics and customized reweighting strategy are selected, our framework can achieve a wide range of desired learning objectives.
scSiameseClu: A Siamese Clustering Framework for Interpreting single-cell RNA Sequencing Data
May 19, 2025



Abstract:Single-cell RNA sequencing (scRNA-seq) reveals cell heterogeneity, with cell clustering playing a key role in identifying cell types and marker genes. Recent advances, especially graph neural networks (GNNs)-based methods, have significantly improved clustering performance. However, the analysis of scRNA-seq data remains challenging due to noise, sparsity, and high dimensionality. Compounding these challenges, GNNs often suffer from over-smoothing, limiting their ability to capture complex biological information. In response, we propose scSiameseClu, a novel Siamese Clustering framework for interpreting single-cell RNA-seq data, comprising of 3 key steps: (1) Dual Augmentation Module, which applies biologically informed perturbations to the gene expression matrix and cell graph relationships to enhance representation robustness; (2) Siamese Fusion Module, which combines cross-correlation refinement and adaptive information fusion to capture complex cellular relationships while mitigating over-smoothing; and (3) Optimal Transport Clustering, which utilizes Sinkhorn distance to efficiently align cluster assignments with predefined proportions while maintaining balance. Comprehensive evaluations on seven real-world datasets demonstrate that~\methodname~outperforms state-of-the-art methods in single-cell clustering, cell type annotation, and cell type classification, providing a powerful tool for scRNA-seq data interpretation.
Deep Cut-informed Graph Embedding and Clustering
Mar 09, 2025Abstract:Graph clustering aims to divide the graph into different clusters. The recently emerging deep graph clustering approaches are largely built on graph neural networks (GNN). However, GNN is designed for general graph encoding and there is a common issue of representation collapse in existing GNN-based deep graph clustering algorithms. We attribute two main reasons for such issue: (i) the inductive bias of GNN models: GNNs tend to generate similar representations for proximal nodes. Since graphs often contain a non-negligible amount of inter-cluster links, the bias results in error message passing and leads to biased clustering; (ii) the clustering guided loss function: most traditional approaches strive to make all samples closer to pre-learned cluster centers, which cause a degenerate solution assigning all data points to a single label thus make all samples and less discriminative. To address these challenges, we investigate graph clustering from a graph cut perspective and propose an innovative and non-GNN-based Deep Cut-informed Graph embedding and Clustering framework, namely DCGC. This framework includes two modules: (i) cut-informed graph encoding; (ii) self-supervised graph clustering via optimal transport. For the encoding module, we derive a cut-informed graph embedding objective to fuse graph structure and attributes by minimizing their joint normalized cut. For the clustering module, we utilize the optimal transport theory to obtain the clustering assignments, which can balance the guidance of proximity to the pre-learned cluster center. With the above two tailored designs, DCGC is more suitable for the graph clustering task, which can effectively alleviate the problem of representation collapse and achieve better performance. We conduct extensive experiments to demonstrate that our method is simple but effective compared with benchmarks.
Towards Trustworthy Federated Learning
Mar 05, 2025



Abstract:This paper develops a comprehensive framework to address three critical trustworthy challenges in federated learning (FL): robustness against Byzantine attacks, fairness, and privacy preservation. To improve the system's defense against Byzantine attacks that send malicious information to bias the system's performance, we develop a Two-sided Norm Based Screening (TNBS) mechanism, which allows the central server to crop the gradients that have the l lowest norms and h highest norms. TNBS functions as a screening tool to filter out potential malicious participants whose gradients are far from the honest ones. To promote egalitarian fairness, we adopt the q-fair federated learning (q-FFL). Furthermore, we adopt a differential privacy-based scheme to prevent raw data at local clients from being inferred by curious parties. Convergence guarantees are provided for the proposed framework under different scenarios. Experimental results on real datasets demonstrate that the proposed framework effectively improves robustness and fairness while managing the trade-off between privacy and accuracy. This work appears to be the first study that experimentally and theoretically addresses fairness, privacy, and robustness in trustworthy FL.
Strategic priorities for transformative progress in advancing biology with proteomics and artificial intelligence
Feb 21, 2025

Abstract:Artificial intelligence (AI) is transforming scientific research, including proteomics. Advances in mass spectrometry (MS)-based proteomics data quality, diversity, and scale, combined with groundbreaking AI techniques, are unlocking new challenges and opportunities in biological discovery. Here, we highlight key areas where AI is driving innovation, from data analysis to new biological insights. These include developing an AI-friendly ecosystem for proteomics data generation, sharing, and analysis; improving peptide and protein identification and quantification; characterizing protein-protein interactions and protein complexes; advancing spatial and perturbation proteomics; integrating multi-omics data; and ultimately enabling AI-empowered virtual cells.
Approximating Discrimination Within Models When Faced With Several Non-Binary Sensitive Attributes
Aug 12, 2024Abstract:Discrimination mitigation with machine learning (ML) models could be complicated because multiple factors may interweave with each other including hierarchically and historically. Yet few existing fairness measures are able to capture the discrimination level within ML models in the face of multiple sensitive attributes. To bridge this gap, we propose a fairness measure based on distances between sets from a manifold perspective, named as 'harmonic fairness measure via manifolds (HFM)' with two optional versions, which can deal with a fine-grained discrimination evaluation for several sensitive attributes of multiple values. To accelerate the computation of distances of sets, we further propose two approximation algorithms named 'Approximation of distance between sets for one sensitive attribute with multiple values (ApproxDist)' and 'Approximation of extended distance between sets for several sensitive attributes with multiple values (ExtendDist)' to respectively resolve bias evaluation of one single sensitive attribute with multiple values and that of several sensitive attributes with multiple values. Moreover, we provide an algorithmic effectiveness analysis for ApproxDist under certain assumptions to explain how well it could work. The empirical results demonstrate that our proposed fairness measure HFM is valid and approximation algorithms (i.e., ApproxDist and ExtendDist) are effective and efficient.
scCDCG: Efficient Deep Structural Clustering for single-cell RNA-seq via Deep Cut-informed Graph Embedding
Apr 09, 2024



Abstract:Single-cell RNA sequencing (scRNA-seq) is essential for unraveling cellular heterogeneity and diversity, offering invaluable insights for bioinformatics advancements. Despite its potential, traditional clustering methods in scRNA-seq data analysis often neglect the structural information embedded in gene expression profiles, crucial for understanding cellular correlations and dependencies. Existing strategies, including graph neural networks, face challenges in handling the inefficiency due to scRNA-seq data's intrinsic high-dimension and high-sparsity. Addressing these limitations, we introduce scCDCG (single-cell RNA-seq Clustering via Deep Cut-informed Graph), a novel framework designed for efficient and accurate clustering of scRNA-seq data that simultaneously utilizes intercellular high-order structural information. scCDCG comprises three main components: (i) A graph embedding module utilizing deep cut-informed techniques, which effectively captures intercellular high-order structural information, overcoming the over-smoothing and inefficiency issues prevalent in prior graph neural network methods. (ii) A self-supervised learning module guided by optimal transport, tailored to accommodate the unique complexities of scRNA-seq data, specifically its high-dimension and high-sparsity. (iii) An autoencoder-based feature learning module that simplifies model complexity through effective dimension reduction and feature extraction. Our extensive experiments on 6 datasets demonstrate scCDCG's superior performance and efficiency compared to 7 established models, underscoring scCDCG's potential as a transformative tool in scRNA-seq data analysis. Our code is available at: https://github.com/XPgogogo/scCDCG.
Robust Distributed Learning Against Both Distributional Shifts and Byzantine Attacks
Oct 29, 2022



Abstract:In distributed learning systems, robustness issues may arise from two sources. On one hand, due to distributional shifts between training data and test data, the trained model could exhibit poor out-of-sample performance. On the other hand, a portion of working nodes might be subject to byzantine attacks which could invalidate the learning result. Existing works mostly deal with these two issues separately. In this paper, we propose a new algorithm that equips distributed learning with robustness measures against both distributional shifts and byzantine attacks. Our algorithm is built on recent advances in distributionally robust optimization as well as norm-based screening (NBS), a robust aggregation scheme against byzantine attacks. We provide convergence proofs in three cases of the learning model being nonconvex, convex, and strongly convex for the proposed algorithm, shedding light on its convergence behaviors and endurability against byzantine attacks. In particular, we deduce that any algorithm employing NBS (including ours) cannot converge when the percentage of byzantine nodes is 1/3 or higher, instead of 1/2, which is the common belief in current literature. The experimental results demonstrate the effectiveness of our algorithm against both robustness issues. To the best of our knowledge, this is the first work to address distributional shifts and byzantine attacks simultaneously.
QC-ODKLA: Quantized and Communication-Censored Online Decentralized Kernel Learning via Linearized ADMM
Aug 04, 2022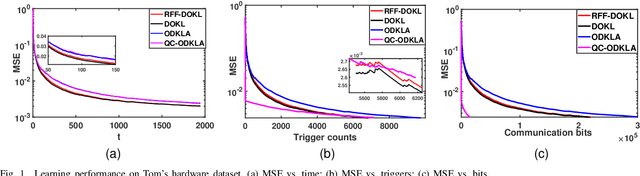
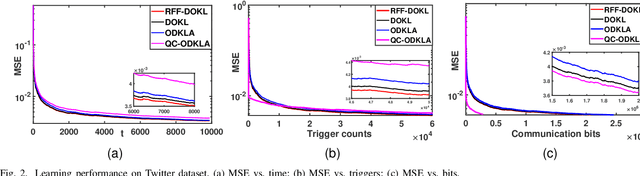
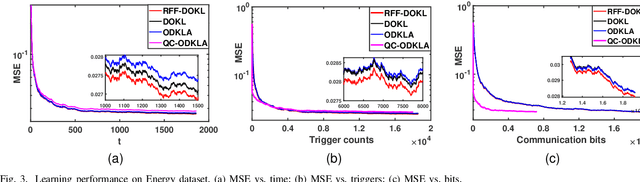
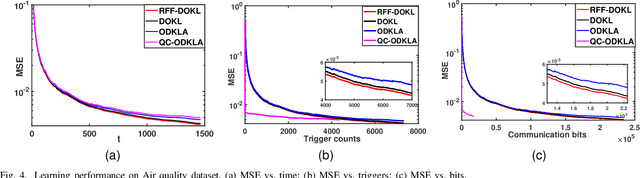
Abstract:This paper focuses on online kernel learning over a decentralized network. Each agent in the network receives continuous streaming data locally and works collaboratively to learn a nonlinear prediction function that is globally optimal in the reproducing kernel Hilbert space with respect to the total instantaneous costs of all agents. In order to circumvent the curse of dimensionality issue in traditional online kernel learning, we utilize random feature (RF) mapping to convert the non-parametric kernel learning problem into a fixed-length parametric one in the RF space. We then propose a novel learning framework named Online Decentralized Kernel learning via Linearized ADMM (ODKLA) to efficiently solve the online decentralized kernel learning problem. To further improve the communication efficiency, we add the quantization and censoring strategies in the communication stage and develop the Quantized and Communication-censored ODKLA (QC-ODKLA) algorithm. We theoretically prove that both ODKLA and QC-ODKLA can achieve the optimal sublinear regret $\mathcal{O}(\sqrt{T})$ over $T$ time slots. Through numerical experiments, we evaluate the learning effectiveness, communication, and computation efficiencies of the proposed methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge