Mo Zhang
Meta-Cognitive Reinforcement Learning with Self-Doubt and Recovery
Jan 28, 2026Abstract:Robust reinforcement learning methods typically focus on suppressing unreliable experiences or corrupted rewards, but they lack the ability to reason about the reliability of their own learning process. As a result, such methods often either overreact to noise by becoming overly conservative or fail catastrophically when uncertainty accumulates. In this work, we propose a meta-cognitive reinforcement learning framework that enables an agent to assess, regulate, and recover its learning behavior based on internally estimated reliability signals. The proposed method introduces a meta-trust variable driven by Value Prediction Error Stability (VPES), which modulates learning dynamics via fail-safe regulation and gradual trust recovery. Experiments on continuous-control benchmarks with reward corruption demonstrate that recovery-enabled meta-cognitive control achieves higher average returns and significantly reduces late-stage training failures compared to strong robustness baselines.
Channel-Independent Federated Traffic Prediction
Aug 06, 2025

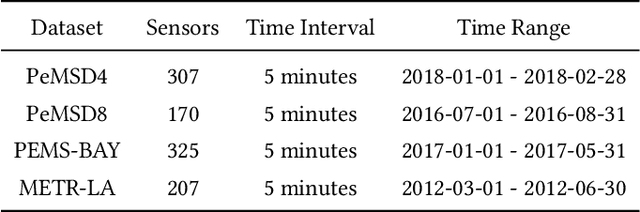

Abstract:In recent years, traffic prediction has achieved remarkable success and has become an integral component of intelligent transportation systems. However, traffic data is typically distributed among multiple data owners, and privacy constraints prevent the direct utilization of these isolated datasets for traffic prediction. Most existing federated traffic prediction methods focus on designing communication mechanisms that allow models to leverage information from other clients in order to improve prediction accuracy. Unfortunately, such approaches often incur substantial communication overhead, and the resulting transmission delays significantly slow down the training process. As the volume of traffic data continues to grow, this issue becomes increasingly critical, making the resource consumption of current methods unsustainable. To address this challenge, we propose a novel variable relationship modeling paradigm for federated traffic prediction, termed the Channel-Independent Paradigm(CIP). Unlike traditional approaches, CIP eliminates the need for inter-client communication by enabling each node to perform efficient and accurate predictions using only local information. Based on the CIP, we further develop Fed-CI, an efficient federated learning framework, allowing each client to process its own data independently while effectively mitigating the information loss caused by the lack of direct data sharing among clients. Fed-CI significantly reduces communication overhead, accelerates the training process, and achieves state-of-the-art performance while complying with privacy regulations. Extensive experiments on multiple real-world datasets demonstrate that Fed-CI consistently outperforms existing methods across all datasets and federated settings. It achieves improvements of 8%, 14%, and 16% in RMSE, MAE, and MAPE, respectively, while also substantially reducing communication costs.
BEFD: Boundary Enhancement and Feature Denoising for Vessel Segmentation
Apr 08, 2021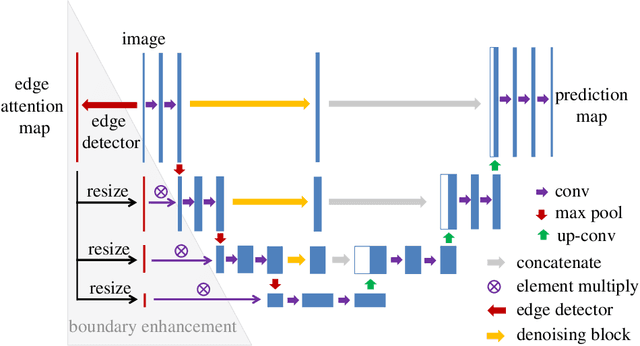
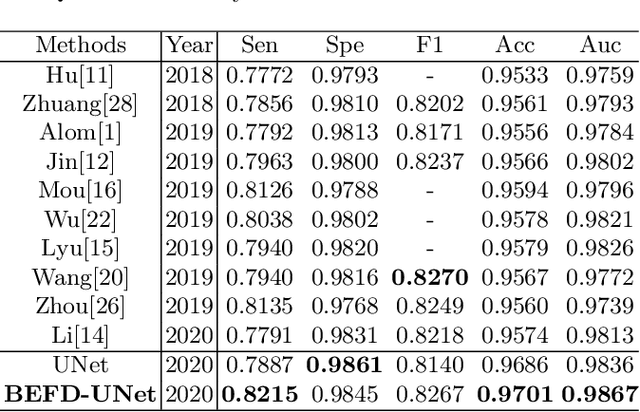
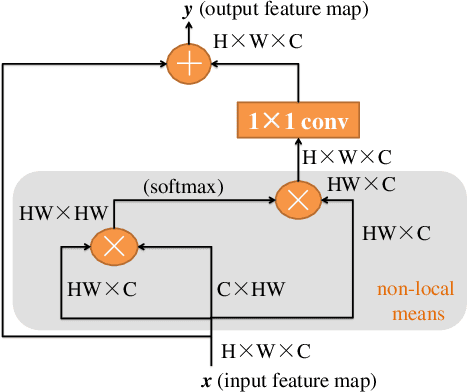
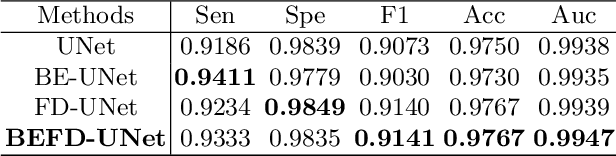
Abstract:Blood vessel segmentation is crucial for many diagnostic and research applications. In recent years, CNN-based models have leaded to breakthroughs in the task of segmentation, however, such methods usually lose high-frequency information like object boundaries and subtle structures, which are vital to vessel segmentation. To tackle this issue, we propose Boundary Enhancement and Feature Denoising (BEFD) module to facilitate the network ability of extracting boundary information in semantic segmentation, which can be integrated into arbitrary encoder-decoder architecture in an end-to-end way. By introducing Sobel edge detector, the network is able to acquire additional edge prior, thus enhancing boundary in an unsupervised manner for medical image segmentation. In addition, we also utilize a denoising block to reduce the noise hidden in the low-level features. Experimental results on retinal vessel dataset and angiocarpy dataset demonstrate the superior performance of the new BEFD module.
MS-GWNN:multi-scale graph wavelet neural network for breast cancer diagnosis
Dec 29, 2020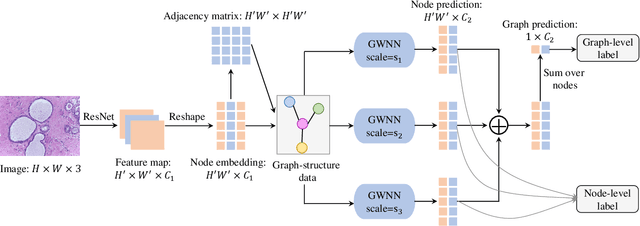
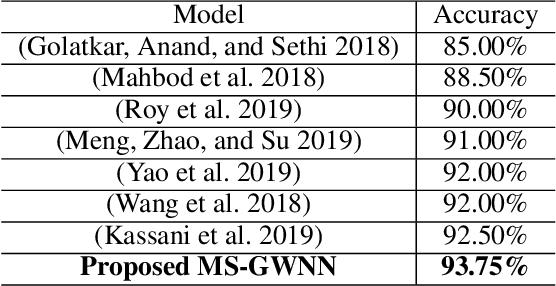
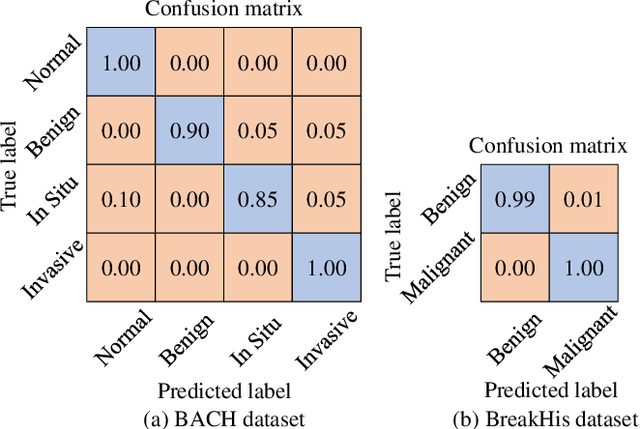
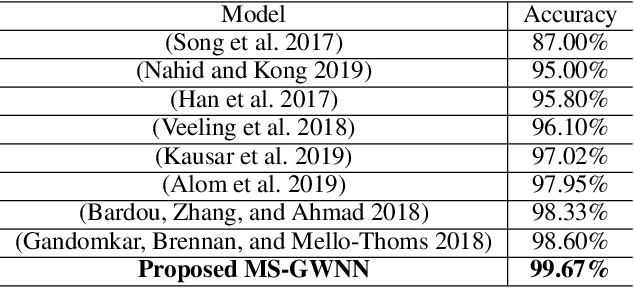
Abstract:Breast cancer is one of the most common cancers in women worldwide, and early detection can significantly reduce the mortality rate of breast cancer. It is crucial to take multi-scale information of tissue structure into account in the detection of breast cancer. And thus, it is the key to design an accurate computer-aided detection (CAD) system to capture multi-scale contextual features in a cancerous tissue. In this work, we present a novel graph convolutional neural network for histopathological image classification of breast cancer. The new method, named multi-scale graph wavelet neural network (MS-GWNN), leverages the localization property of spectral graph wavelet to perform multi-scale analysis. By aggregating features at different scales, MS-GWNN can encode the multi-scale contextual interactions in the whole pathological slide. Experimental results on two public datasets demonstrate the superiority of the proposed method. Moreover, through ablation studies, we find that multi-scale analysis has a significant impact on the accuracy of cancer diagnosis.
PGU-net+: Progressive Growing of U-net+ for Automated Cervical Nuclei Segmentation
Nov 12, 2019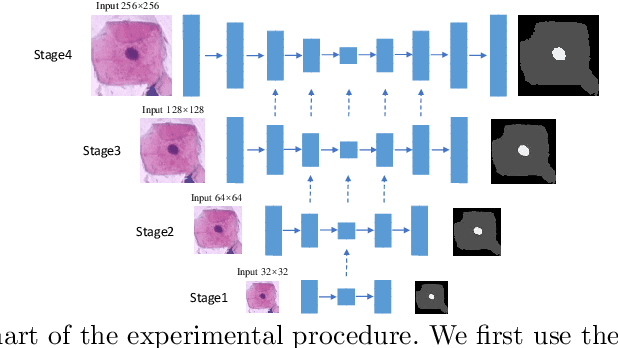
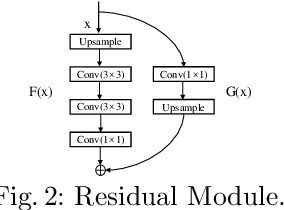


Abstract:Automated cervical nucleus segmentation based on deep learning can effectively improve the quantitative analysis of cervical cancer. However, accurate nuclei segmentation is still challenging. The classic U-net has not achieved satisfactory results on this task, because it mixes the information of different scales that affect each other, which limits the segmentation accuracy of the model. To solve this problem, we propose a progressive growing U-net (PGU-net+) model, which uses two paradigms to extract image features at different scales in a more independent way. First, we add residual modules between different scales of U-net, which enforces the model to learn the approximate shape of the annotation in the coarser scale, and to learn the residual between the annotation and the approximate shape in the finer scale. Second, we start to train the model with the coarsest part and then progressively add finer part to the training until the full model is included. When we train a finer part, we will reduce the learning rate of the previous coarser part, which further ensures that the model independently extracts information from different scales. We conduct several comparative experiments on the Herlev dataset. The experimental results show that the PGU-net+ has superior accuracy than the previous state-of-the-art methods on cervical nuclei segmentation.
Multi-label Detection and Classification of Red Blood Cells in Microscopic Images
Oct 07, 2019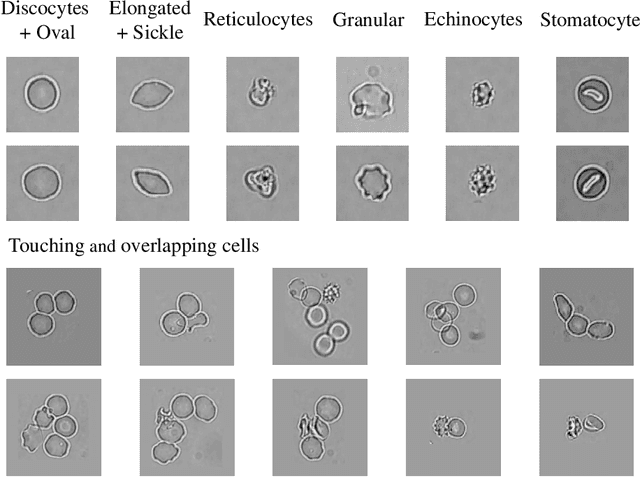
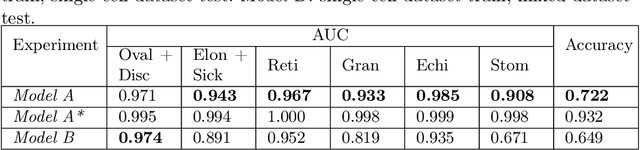
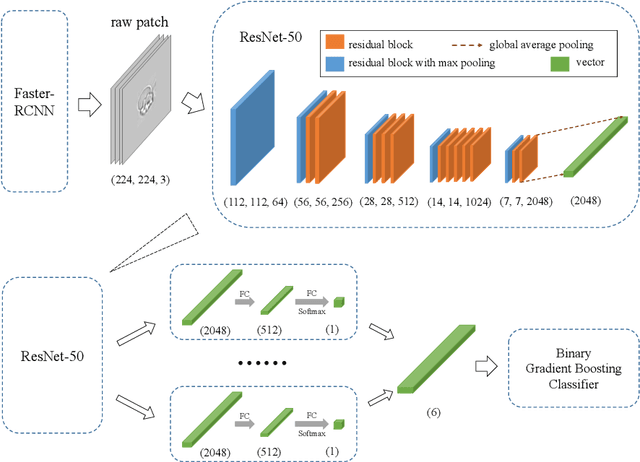
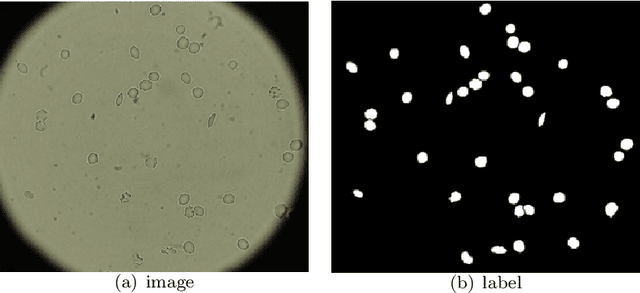
Abstract:Cell detection and cell type classification from biomedical images play an important role for high-throughput imaging and various clinical application. While classification of single cell sample can be performed with standard computer vision and machine learning methods, analysis of multi-label samples (region containing congregating cells) is more challenging, as separation of individual cells can be difficult (e.g. touching cells) or even impossible (e.g. overlapping cells). As multi-instance images are common in analyzing Red Blood Cell (RBC) for Sickle Cell Disease (SCD) diagnosis, we develop and implement a multi-instance cell detection and classification framework to address this challenge. The framework firstly trains a region proposal model based on Region-based Convolutional Network (RCNN) to obtain bounding-boxes of regions potentially containing single or multiple cells from input microscopic images, which are extracted as image patches. High-level image features are then calculated from image patches through a pre-trained Convolutional Neural Network (CNN) with ResNet-50 structure. Using these image features inputs, six networks are then trained to make multi-label prediction of whether a given patch contains cells belonging to a specific cell type. As the six networks are trained with image patches consisting of both individual cells and touching/overlapping cells, they can effectively recognize cell types that are presented in multi-instance image samples. Finally, for the purpose of SCD testing, we train another machine learning classifier to predict whether the given image patch contains abnormal cell type based on outputs from the six networks. Testing result of the proposed framework shows that it can achieve good performance in automatic cell detection and classification.
ASCNet: Adaptive-Scale Convolutional Neural Networks for Multi-Scale Feature Learning
Jul 07, 2019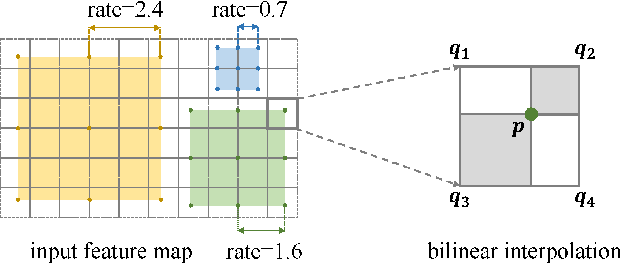
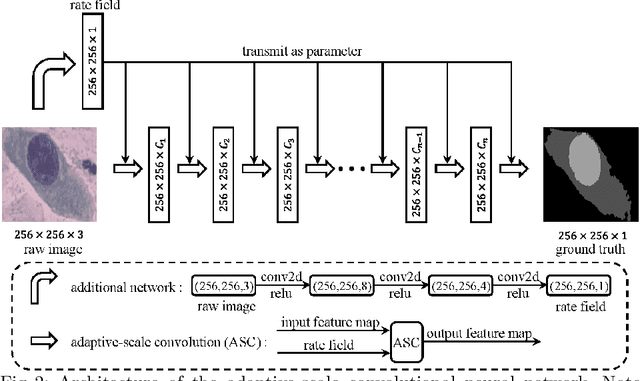
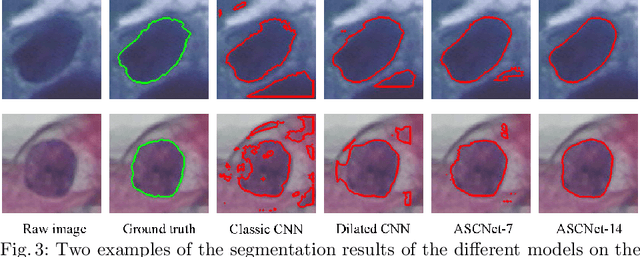
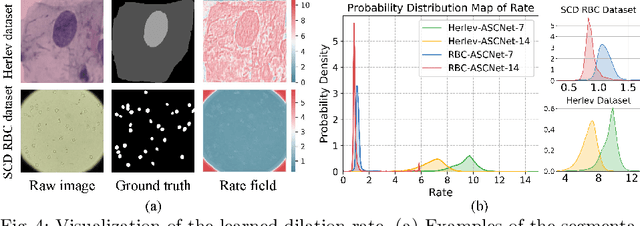
Abstract:Extracting multi-scale information is key to semantic segmentation. However, the classic convolutional neural networks (CNNs) encounter difficulties in achieving multi-scale information extraction: expanding convolutional kernel incurs the high computational cost and using maximum pooling sacrifices image information. The recently developed dilated convolution solves these problems, but with the limitation that the dilation rates are fixed and therefore the receptive field cannot fit for all objects with different sizes in the image. We propose an adaptivescale convolutional neural network (ASCNet), which introduces a 3-layer convolution structure in the end-to-end training, to adaptively learn an appropriate dilation rate for each pixel in the image. Such pixel-level dilation rates produce optimal receptive fields so that the information of objects with different sizes can be extracted at the corresponding scale. We compare the segmentation results using the classic CNN, the dilated CNN and the proposed ASCNet on two types of medical images (The Herlev dataset and SCD RBC dataset). The experimental results show that ASCNet achieves the highest accuracy. Moreover, the automatically generated dilation rates are positively correlated to the sizes of the objects, confirming the effectiveness of the proposed method.
Modeling 4D fMRI Data via Spatio-Temporal Convolutional Neural Networks
Aug 06, 2018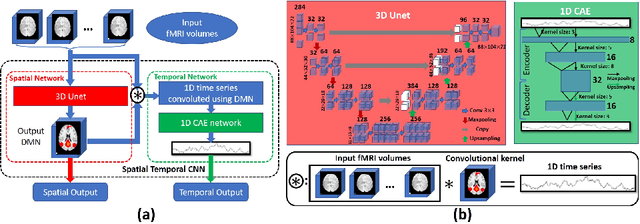
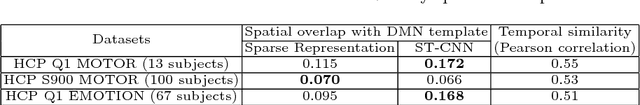
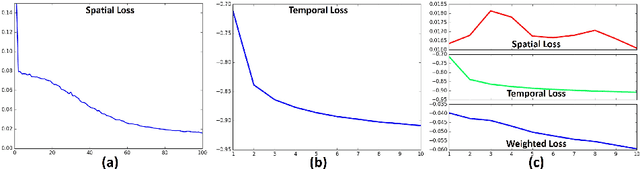
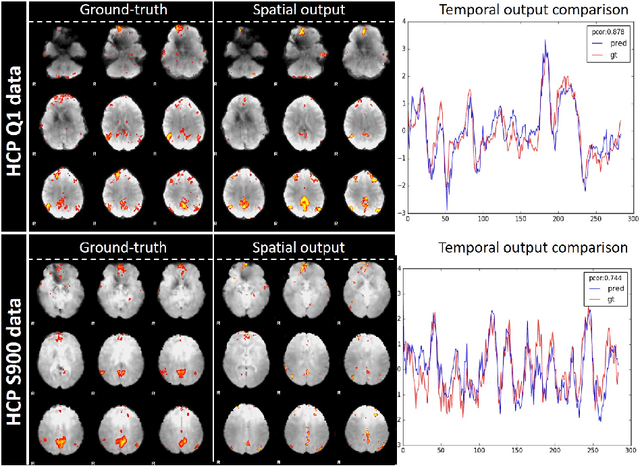
Abstract:Simultaneous modeling of the spatio-temporal variation patterns of brain functional network from 4D fMRI data has been an important yet challenging problem for the field of cognitive neuroscience and medical image analysis. Inspired by the recent success in applying deep learning for functional brain decoding and encoding, in this work we propose a spatio-temporal convolutional neural network (ST-CNN)to jointly learn the spatial and temporal patterns of targeted network from the training data and perform automatic, pin-pointing functional network identification. The proposed ST-CNN is evaluated by the task of identifying the Default Mode Network (DMN) from fMRI data. Results show that while the framework is only trained on one fMRI dataset,it has the sufficient generalizability to identify the DMN from different populations of data as well as different cognitive tasks. Further investigation into the results show that the superior performance of ST-CNN is driven by the jointly-learning scheme, which capture the intrinsic relationship between the spatial and temporal characteristic of DMN and ensures the accurate identification.
Image Segmentation and Classification for Sickle Cell Disease using Deformable U-Net
Oct 29, 2017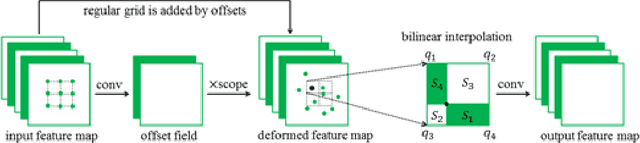
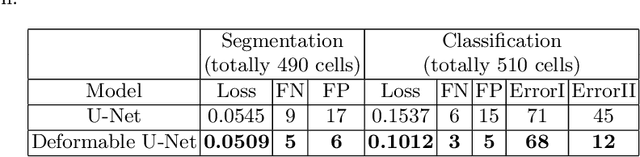
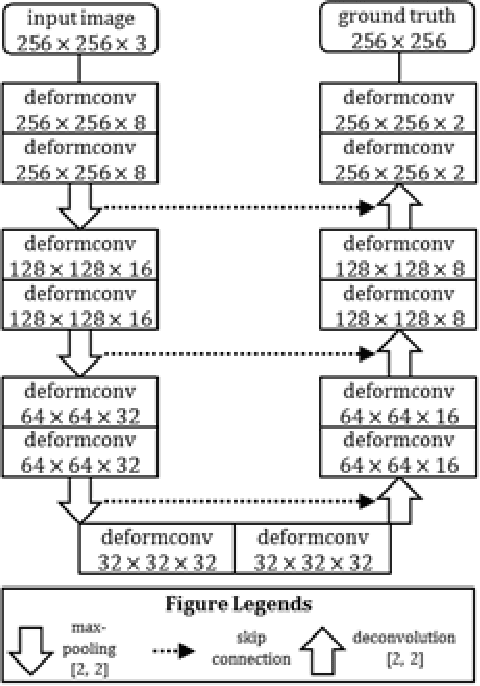
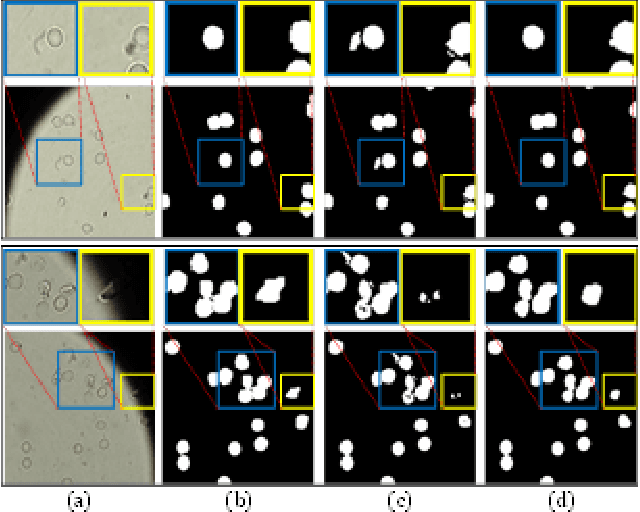
Abstract:Reliable cell segmentation and classification from biomedical images is a crucial step for both scientific research and clinical practice. A major challenge for more robust segmentation and classification methods is the large variations in the size, shape and viewpoint of the cells, combining with the low image quality caused by noise and artifacts. To address this issue, in this work we propose a learning-based, simultaneous cell segmentation and classification method based on the deep U-Net structure with deformable convolution layers. The U-Net architecture for deep learning has been shown to offer a precise localization for image semantic segmentation. Moreover, deformable convolution layer enables the free form deformation of the feature learning process, thus makes the whole network more robust to various cell morphologies and image settings. The proposed method is tested on microscopic red blood cell images from patients with sickle cell disease. The results show that U-Net with deformable convolution achieves the highest accuracy for segmentation and classification, comparing with original U-Net structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge