Maximilian Baust
Spherical Fourier Neural Operators: Learning Stable Dynamics on the Sphere
Jun 06, 2023



Abstract:Fourier Neural Operators (FNOs) have proven to be an efficient and effective method for resolution-independent operator learning in a broad variety of application areas across scientific machine learning. A key reason for their success is their ability to accurately model long-range dependencies in spatio-temporal data by learning global convolutions in a computationally efficient manner. To this end, FNOs rely on the discrete Fourier transform (DFT), however, DFTs cause visual and spectral artifacts as well as pronounced dissipation when learning operators in spherical coordinates since they incorrectly assume a flat geometry. To overcome this limitation, we generalize FNOs on the sphere, introducing Spherical FNOs (SFNOs) for learning operators on spherical geometries. We apply SFNOs to forecasting atmospheric dynamics, and demonstrate stable auto\-regressive rollouts for a year of simulated time (1,460 steps), while retaining physically plausible dynamics. The SFNO has important implications for machine learning-based simulation of climate dynamics that could eventually help accelerate our response to climate change.
The Future of Digital Health with Federated Learning
Mar 18, 2020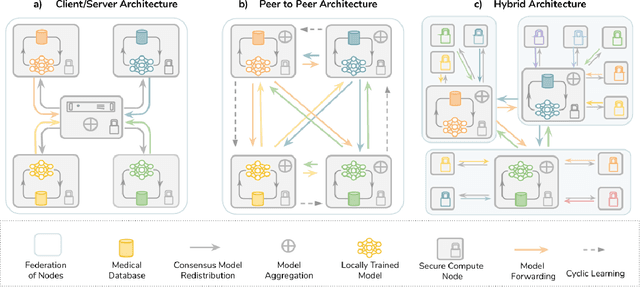
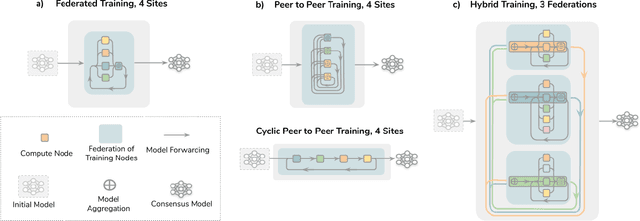
Abstract:Data-driven Machine Learning has emerged as a promising approach for building accurate and robust statistical models from medical data, which is collected in huge volumes by modern healthcare systems. Existing medical data is not fully exploited by ML primarily because it sits in data silos and privacy concerns restrict access to this data. However, without access to sufficient data, ML will be prevented from reaching its full potential and, ultimately, from making the transition from research to clinical practice. This paper considers key factors contributing to this issue, explores how Federated Learning (FL) may provide a solution for the future of digital health and highlights the challenges and considerations that need to be addressed.
Privacy-preserving Federated Brain Tumour Segmentation
Oct 02, 2019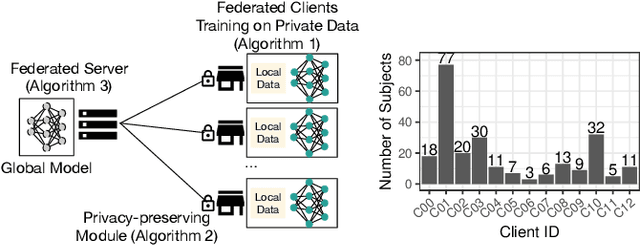
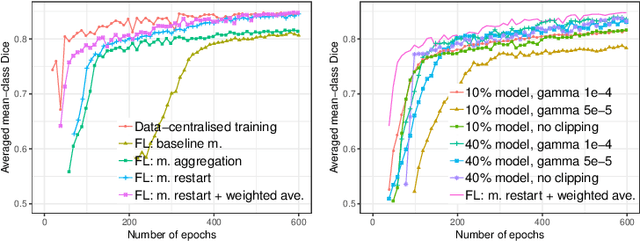
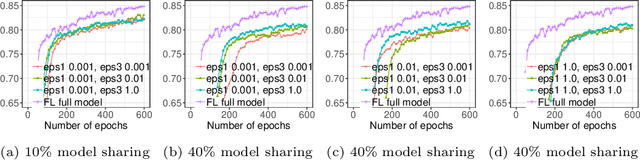
Abstract:Due to medical data privacy regulations, it is often infeasible to collect and share patient data in a centralised data lake. This poses challenges for training machine learning algorithms, such as deep convolutional networks, which often require large numbers of diverse training examples. Federated learning sidesteps this difficulty by bringing code to the patient data owners and only sharing intermediate model training updates among them. Although a high-accuracy model could be achieved by appropriately aggregating these model updates, the model shared could indirectly leak the local training examples. In this paper, we investigate the feasibility of applying differential-privacy techniques to protect the patient data in a federated learning setup. We implement and evaluate practical federated learning systems for brain tumour segmentation on the BraTS dataset. The experimental results show that there is a trade-off between model performance and privacy protection costs.
Redefining Ultrasound Compounding: Computational Sonography
Nov 05, 2018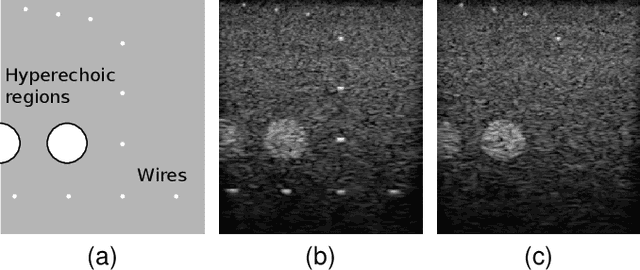
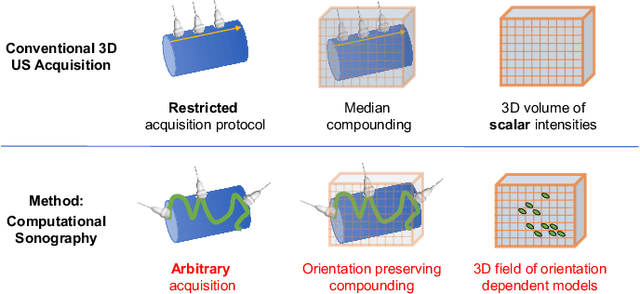
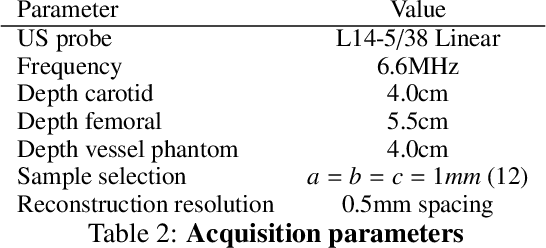
Abstract:Freehand three-dimensional ultrasound (3D-US) has gained considerable interest in research, but even today suffers from its high inter-operator variability in clinical practice. The high variability mainly arises from tracking inaccuracies as well as the directionality of the ultrasound data, being neglected in most of today's reconstruction methods. By providing a novel paradigm for the acquisition and reconstruction of tracked freehand 3D ultrasound, this work presents the concept of Computational Sonography (CS) to model the directionality of ultrasound information. CS preserves the directionality of the acquired data, and allows for its exploitation by computational algorithms. In this regard, we propose a set of mathematical models to represent 3D-US data, inspired by the physics of ultrasound imaging. We compare different models of Computational Sonography to classical scalar compounding for freehand acquisitions, providing both an improved preservation of US directionality as well as improved image quality in 3D. The novel concept is evaluated for a set of phantom datasets, as well as for in-vivo acquisitions of muscoloskeletal and vascular applications.
BACH: Grand Challenge on Breast Cancer Histology Images
Aug 13, 2018
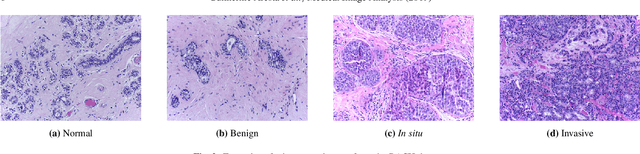
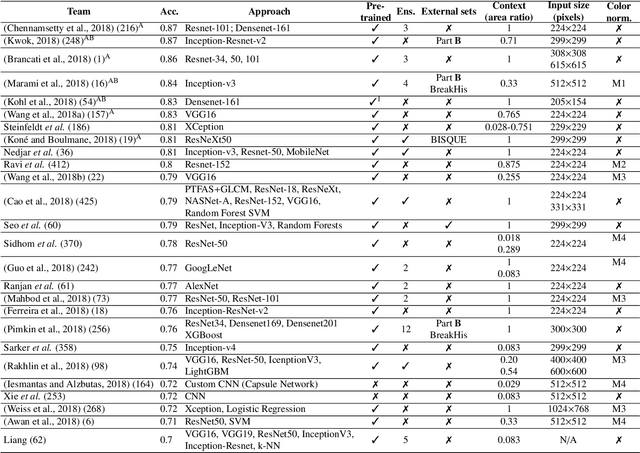
Abstract:Breast cancer is the most common invasive cancer in women, affecting more than 10% of women worldwide. Microscopic analysis of a biopsy remains one of the most important methods to diagnose the type of breast cancer. This requires specialized analysis by pathologists, in a task that i) is highly time- and cost-consuming and ii) often leads to nonconsensual results. The relevance and potential of automatic classification algorithms using hematoxylin-eosin stained histopathological images has already been demonstrated, but the reported results are still sub-optimal for clinical use. With the goal of advancing the state-of-the-art in automatic classification, the Grand Challenge on BreAst Cancer Histology images (BACH) was organized in conjunction with the 15th International Conference on Image Analysis and Recognition (ICIAR 2018). A large annotated dataset, composed of both microscopy and whole-slide images, was specifically compiled and made publicly available for the BACH challenge. Following a positive response from the scientific community, a total of 64 submissions, out of 677 registrations, effectively entered the competition. From the submitted algorithms it was possible to push forward the state-of-the-art in terms of accuracy (87%) in automatic classification of breast cancer with histopathological images. Convolutional neuronal networks were the most successful methodology in the BACH challenge. Detailed analysis of the collective results allowed the identification of remaining challenges in the field and recommendations for future developments. The BACH dataset remains publically available as to promote further improvements to the field of automatic classification in digital pathology.
Initialize globally before acting locally: Enabling Landmark-free 3D US to MRI Registration
Jun 12, 2018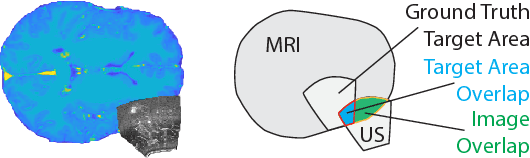
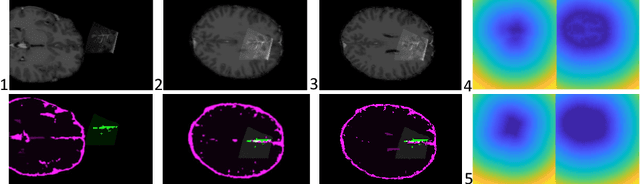
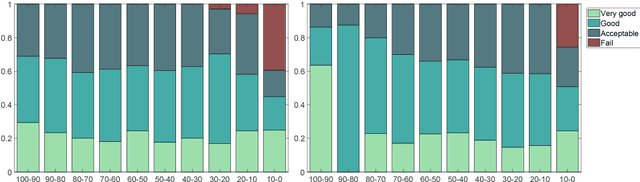
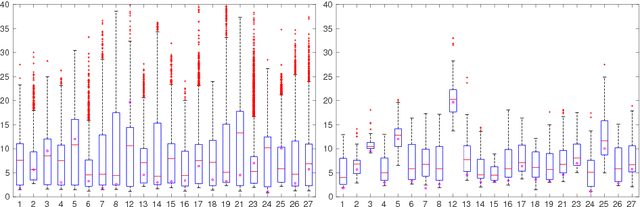
Abstract:Registration of partial-view 3D US volumes with MRI data is influenced by initialization. The standard of practice is using extrinsic or intrinsic landmarks, which can be very tedious to obtain. To overcome the limitations of registration initialization, we present a novel approach that is based on Euclidean distance maps derived from easily obtainable coarse segmentations. We evaluate our approach quantitatively on the publicly available RESECT dataset and show that it is robust regarding overlap of target area and initial position. Furthermore, our method provides initializations that are suitable for state-of-the-art nonlinear, deformable image registration algorithm's capture ranges.
CFCM: Segmentation via Coarse to Fine Context Memory
Jun 04, 2018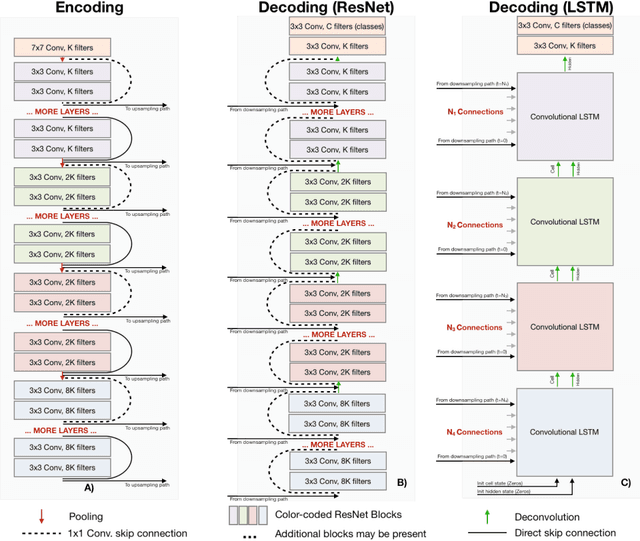
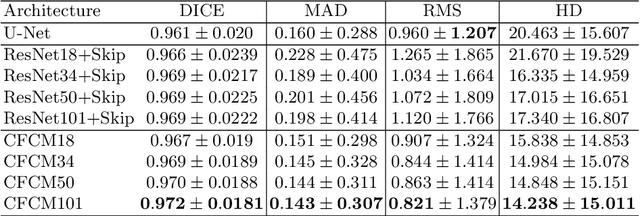
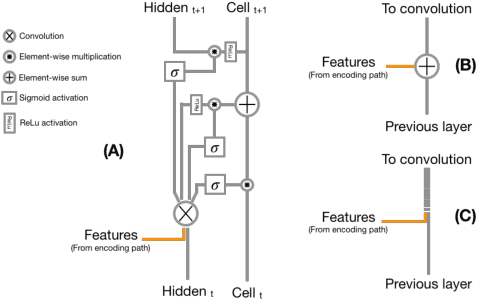
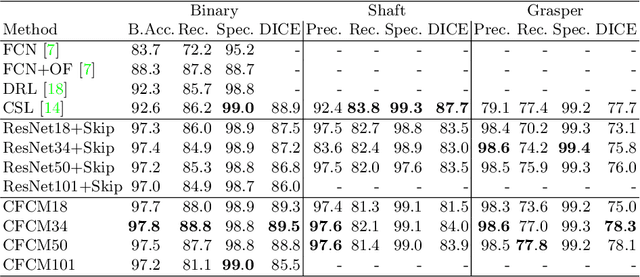
Abstract:Recent neural-network-based architectures for image segmentation make extensive usage of feature forwarding mechanisms to integrate information from multiple scales. Although yielding good results, even deeper architectures and alternative methods for feature fusion at different resolutions have been scarcely investigated for medical applications. In this work we propose to implement segmentation via an encoder-decoder architecture which differs from any other previously published method since (i) it employs a very deep architecture based on residual learning and (ii) combines features via a convolutional Long Short Term Memory (LSTM), instead of concatenation or summation. The intuition is that the memory mechanism implemented by LSTMs can better integrate features from different scales through a coarse-to-fine strategy; hence the name Coarse-to-Fine Context Memory (CFCM). We demonstrate the remarkable advantages of this approach on two datasets: the Montgomery county lung segmentation dataset, and the EndoVis 2015 challenge dataset for surgical instrument segmentation.
Understanding Regularization to Visualize Convolutional Neural Networks
Apr 20, 2018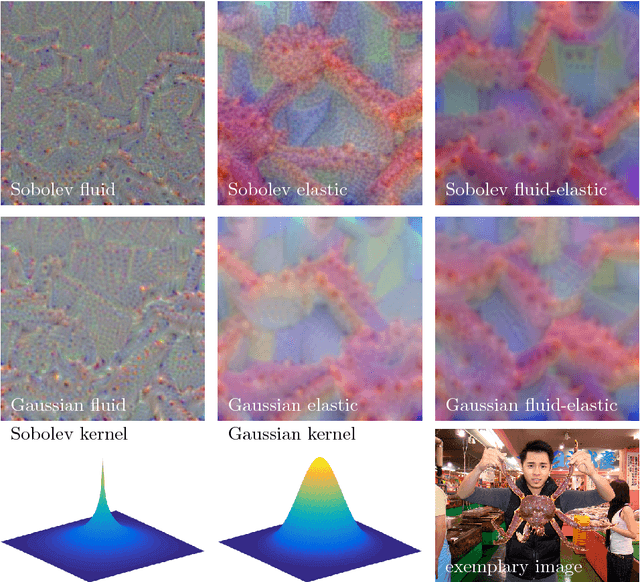
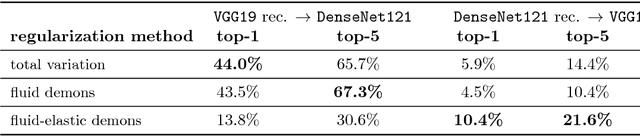
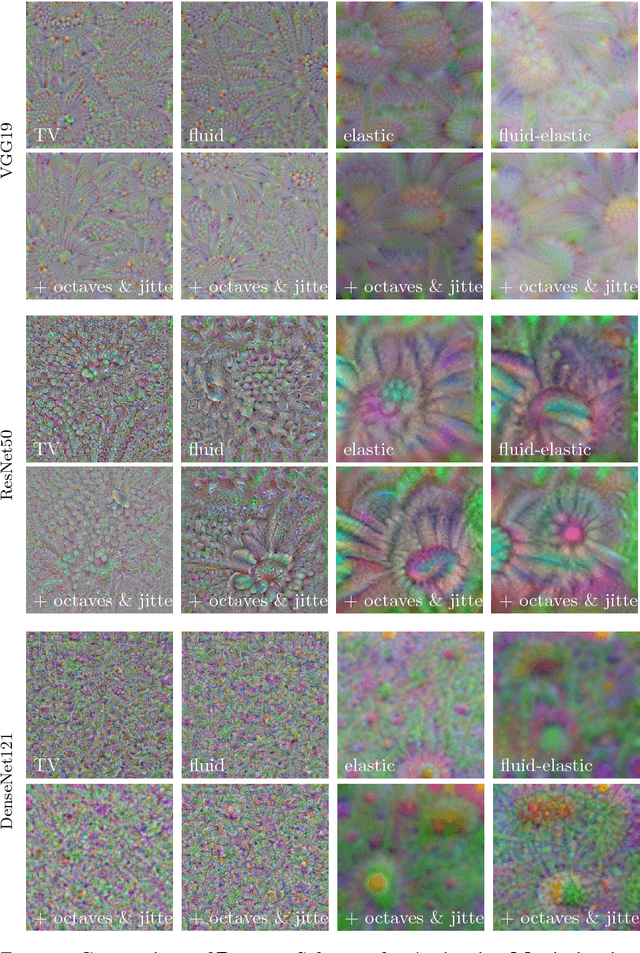
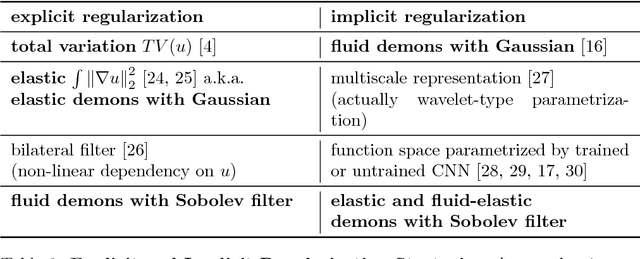
Abstract:Variational methods for revealing visual concepts learned by convolutional neural networks have gained significant attention during the last years. Being based on noisy gradients obtained via back-propagation such methods require the application of regularization strategies. We present a mathematical framework unifying previously employed regularization methods. Within this framework, we propose a novel technique based on Sobolev gradients which can be implemented via convolutions and does not require specialized numerical treatment, such as total variation regularization. The experiments performed on feature inversion and activation maximization demonstrate the benefit of a unified approach to regularization, such as sharper reconstructions via the proposed Sobolev filters and a better control over reconstructed scales.
Assessment of Breast Cancer Histology using Densely Connected Convolutional Networks
Apr 09, 2018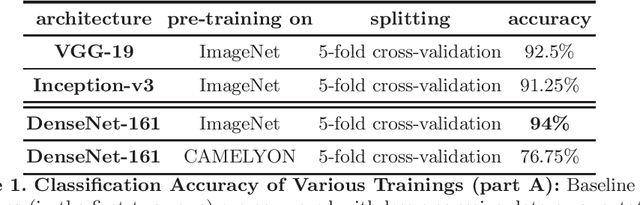

Abstract:Breast cancer is the most frequently diagnosed cancer and leading cause of cancer-related death among females worldwide. In this article, we investigate the applicability of densely connected convolutional neural networks to the problems of histology image classification and whole slide image segmentation in the area of computer-aided diagnoses for breast cancer. To this end, we study various approaches for transfer learning and apply them to the data set from the 2018 grand challenge on breast cancer histology images (BACH).
Learning in an Uncertain World: Representing Ambiguity Through Multiple Hypotheses
Aug 22, 2017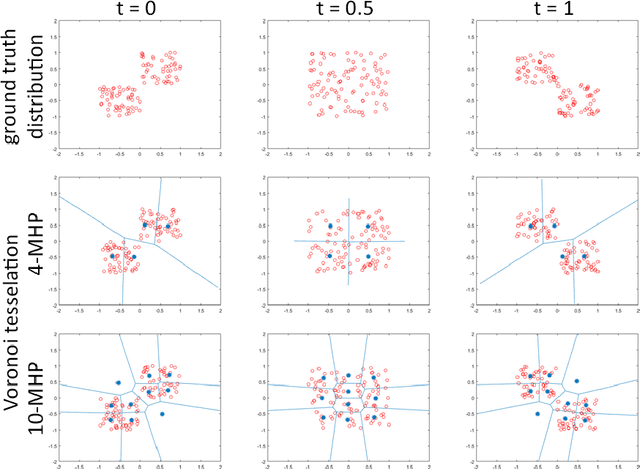
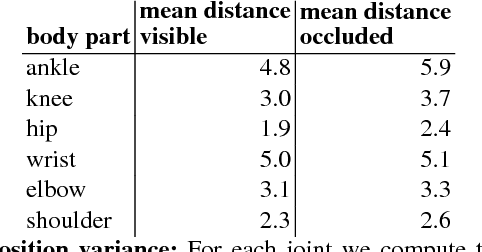

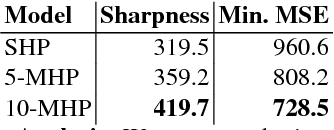
Abstract:Many prediction tasks contain uncertainty. In some cases, uncertainty is inherent in the task itself. In future prediction, for example, many distinct outcomes are equally valid. In other cases, uncertainty arises from the way data is labeled. For example, in object detection, many objects of interest often go unlabeled, and in human pose estimation, occluded joints are often labeled with ambiguous values. In this work we focus on a principled approach for handling such scenarios. In particular, we propose a framework for reformulating existing single-prediction models as multiple hypothesis prediction (MHP) models and an associated meta loss and optimization procedure to train them. To demonstrate our approach, we consider four diverse applications: human pose estimation, future prediction, image classification and segmentation. We find that MHP models outperform their single-hypothesis counterparts in all cases, and that MHP models simultaneously expose valuable insights into the variability of predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge