Varghese Alex
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020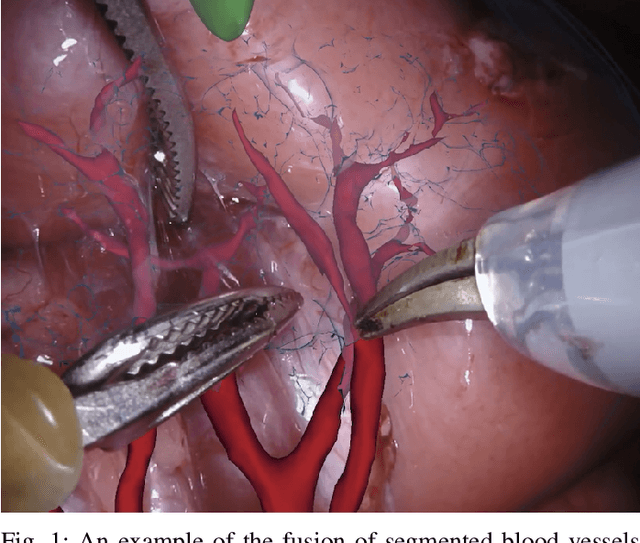
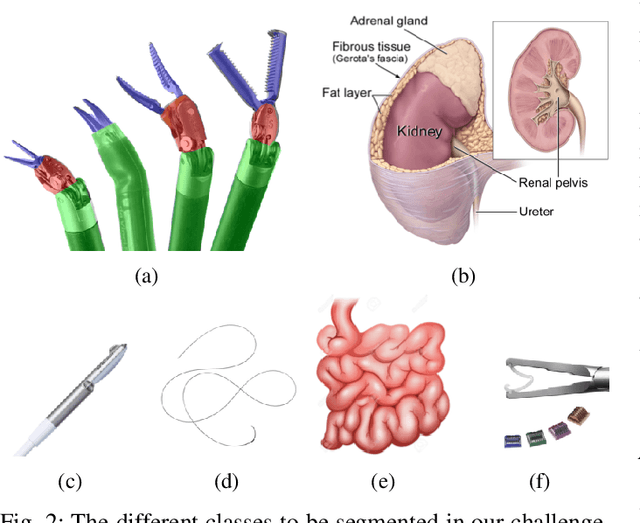
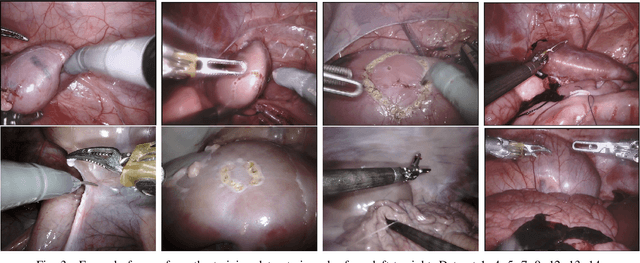
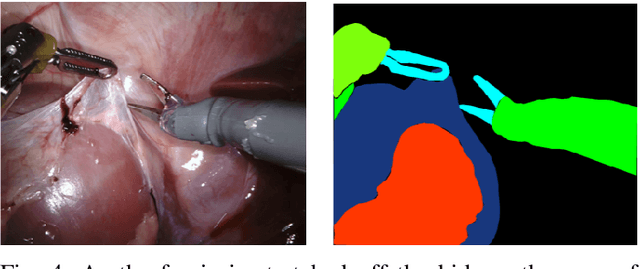
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
Enhanced Optic Disk and Cup Segmentation with Glaucoma Screening from Fundus Images using Position encoded CNNs
Sep 14, 2018
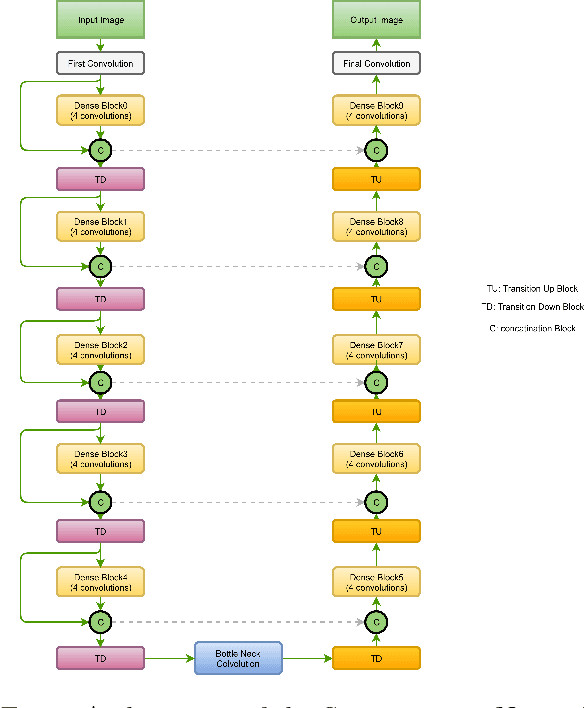
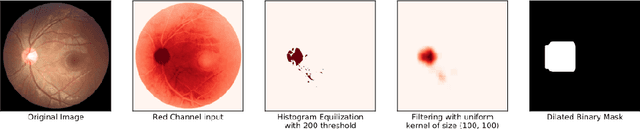
Abstract:In this manuscript, we present a robust method for glaucoma screening from fundus images using an ensemble of convolutional neural networks (CNNs). The pipeline comprises of first segmenting the optic disk and optic cup from the fundus image, then extracting a patch centered around the optic disk and subsequently feeding to the classification network to differentiate the image as diseased or healthy. In the segmentation network, apart from the image, we make use of spatial co-ordinate (X \& Y) space so as to learn the structure of interest better. The classification network is composed of a DenseNet201 and a ResNet18 which were pre-trained on a large cohort of natural images. On the REFUGE validation data (n=400), the segmentation network achieved a dice score of 0.88 and 0.64 for optic disc and optic cup respectively. For the tasking differentiating images affected with glaucoma from healthy images, the area under the ROC curve was observed to be 0.85.
Ensemble of Convolutional Neural Networks for Automatic Grading of Diabetic Retinopathy and Macular Edema
Sep 12, 2018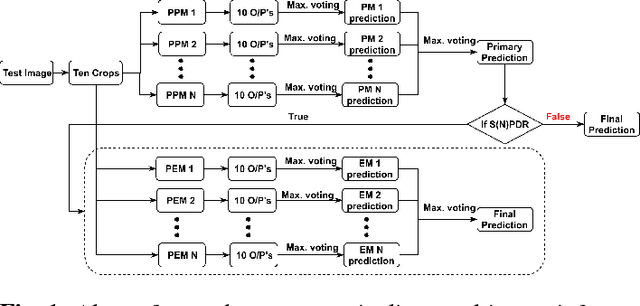
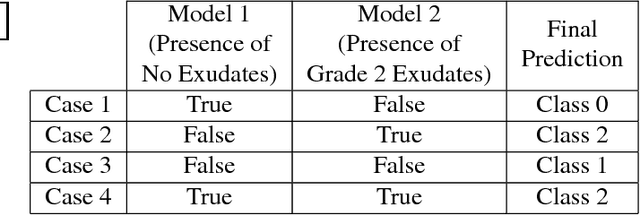
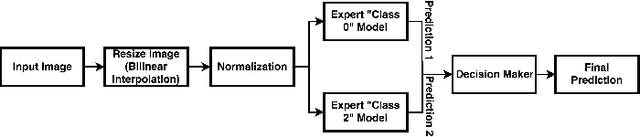
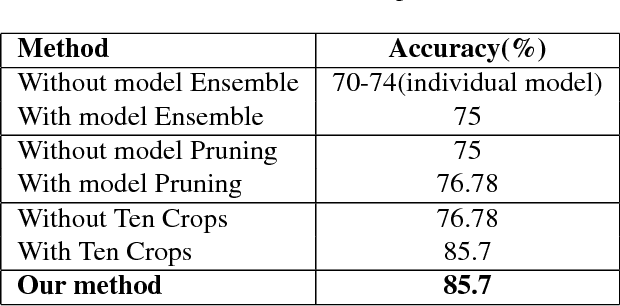
Abstract:In this manuscript, we automate the procedure of grading of diabetic retinopathy and macular edema from fundus images using an ensemble of convolutional neural networks. The availability of limited amount of labeled data to perform supervised learning was circumvented by using transfer learning approach. The models in the ensemble were pre-trained on a large dataset comprising natural images and were later fine-tuned with the limited data for the task of choice. For an image, the ensemble of classifiers generate multiple predictions, and a max-voting based approach was utilized to attain the final grade of the anomaly in the image. For the task of grading DR, on the test data (n=56), the ensemble achieved an accuracy of 83.9\%, while for the task for grading macular edema the network achieved an accuracy of 95.45% (n=44).
BACH: Grand Challenge on Breast Cancer Histology Images
Aug 13, 2018
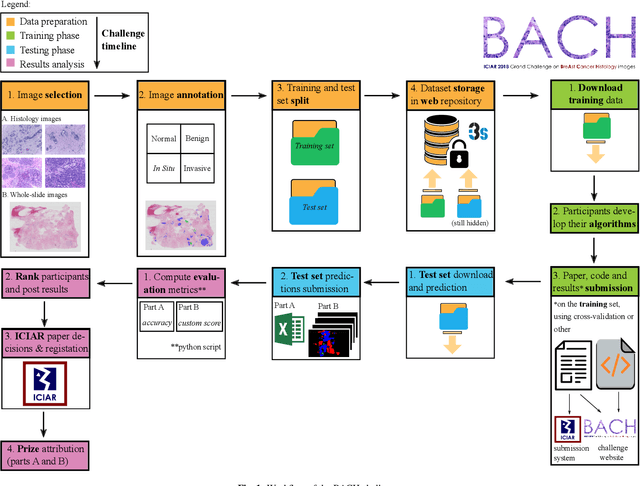
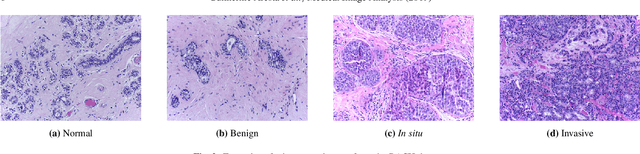
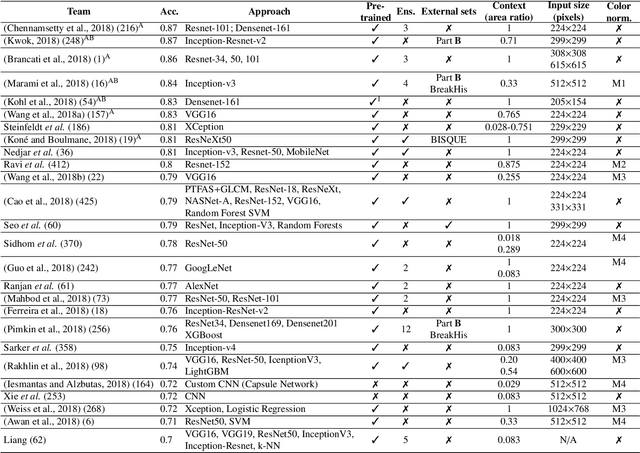
Abstract:Breast cancer is the most common invasive cancer in women, affecting more than 10% of women worldwide. Microscopic analysis of a biopsy remains one of the most important methods to diagnose the type of breast cancer. This requires specialized analysis by pathologists, in a task that i) is highly time- and cost-consuming and ii) often leads to nonconsensual results. The relevance and potential of automatic classification algorithms using hematoxylin-eosin stained histopathological images has already been demonstrated, but the reported results are still sub-optimal for clinical use. With the goal of advancing the state-of-the-art in automatic classification, the Grand Challenge on BreAst Cancer Histology images (BACH) was organized in conjunction with the 15th International Conference on Image Analysis and Recognition (ICIAR 2018). A large annotated dataset, composed of both microscopy and whole-slide images, was specifically compiled and made publicly available for the BACH challenge. Following a positive response from the scientific community, a total of 64 submissions, out of 677 registrations, effectively entered the competition. From the submitted algorithms it was possible to push forward the state-of-the-art in terms of accuracy (87%) in automatic classification of breast cancer with histopathological images. Convolutional neuronal networks were the most successful methodology in the BACH challenge. Detailed analysis of the collective results allowed the identification of remaining challenges in the field and recommendations for future developments. The BACH dataset remains publically available as to promote further improvements to the field of automatic classification in digital pathology.
Automatic Segmentation and Overall Survival Prediction in Gliomas using Fully Convolutional Neural Network and Texture Analysis
Dec 06, 2017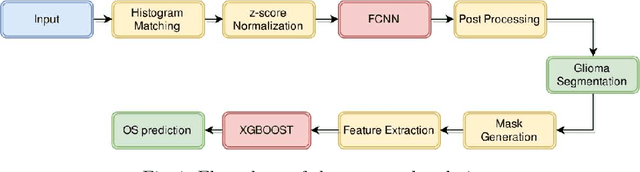
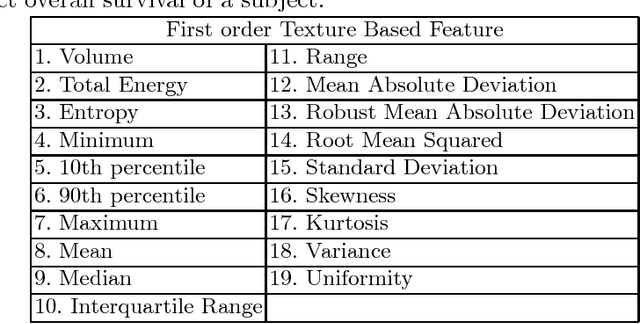
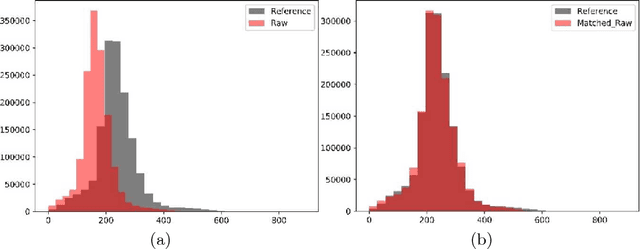
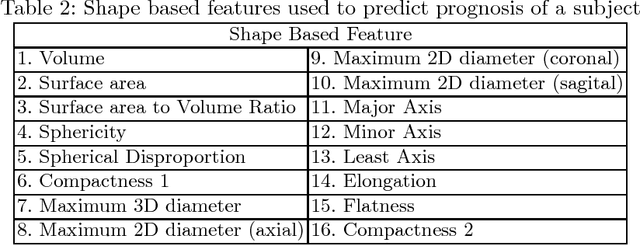
Abstract:In this paper, we use a fully convolutional neural network (FCNN) for the segmentation of gliomas from Magnetic Resonance Images (MRI). A fully automatic, voxel based classification was achieved by training a 23 layer deep FCNN on 2-D slices extracted from patient volumes. The network was trained on slices extracted from 130 patients and validated on 50 patients. For the task of survival prediction, texture and shape based features were extracted from T1 post contrast volume to train an XGBoost regressor. On BraTS 2017 validation set, the proposed scheme achieved a mean whole tumor, tumor core and active dice score of 0.83, 0.69 and 0.69 respectively and an accuracy of 52% for the overall survival prediction.
Semi-supervised Learning using Denoising Autoencoders for Brain Lesion Detection and Segmentation
Jan 10, 2017Abstract:The work presented explores the use of denoising autoencoders (DAE) for brain lesion detection, segmentation and false positive reduction. Stacked denoising autoencoders (SDAE) were pre-trained using a large number of unlabeled patient volumes and fine tuned with patches drawn from a limited number of patients (n=20, 40, 65). The results show negligible loss in performance even when SDAE was fine tuned using 20 patients. Low grade glioma (LGG) segmentation was achieved using a transfer learning approach wherein a network pre-trained with High Grade Glioma (HGG) data was fine tuned using LGG image patches. The weakly supervised SDAE (for HGG) and transfer learning based LGG network were also shown to generalize well and provide good segmentation on unseen BraTS 2013 & BraTS 2015 test data. An unique contribution includes a single layer DAE, referred to as novelty detector(ND). ND was trained to accurately reconstruct non-lesion patches using a mean squared error loss function. The reconstruction error maps of test data were used to identify regions containing lesions. The error maps were shown to assign unique error distributions to various constituents of the glioma, enabling localization. The ND learns the non-lesion brain accurately as it was also shown to provide good segmentation performance on ischemic brain lesions in images from a different database.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge