Leon Y. Cai
Tractography with T1-weighted MRI and associated anatomical constraints on clinical quality diffusion MRI
Mar 27, 2024Abstract:Diffusion MRI (dMRI) streamline tractography, the gold standard for in vivo estimation of brain white matter (WM) pathways, has long been considered indicative of macroscopic relationships with WM microstructure. However, recent advances in tractography demonstrated that convolutional recurrent neural networks (CoRNN) trained with a teacher-student framework have the ability to learn and propagate streamlines directly from T1 and anatomical contexts. Training for this network has previously relied on high-resolution dMRI. In this paper, we generalize the training mechanism to traditional clinical resolution data, which allows generalizability across sensitive and susceptible study populations. We train CoRNN on a small subset of the Baltimore Longitudinal Study of Aging (BLSA), which better resembles clinical protocols. Then, we define a metric, termed the epsilon ball seeding method, to compare T1 tractography and traditional diffusion tractography at the streamline level. Under this metric, T1 tractography generated by CoRNN reproduces diffusion tractography with approximately two millimeters of error.
Nucleus subtype classification using inter-modality learning
Jan 29, 2024Abstract:Understanding the way cells communicate, co-locate, and interrelate is essential to understanding human physiology. Hematoxylin and eosin (H&E) staining is ubiquitously available both for clinical studies and research. The Colon Nucleus Identification and Classification (CoNIC) Challenge has recently innovated on robust artificial intelligence labeling of six cell types on H&E stains of the colon. However, this is a very small fraction of the number of potential cell classification types. Specifically, the CoNIC Challenge is unable to classify epithelial subtypes (progenitor, endocrine, goblet), lymphocyte subtypes (B, helper T, cytotoxic T), or connective subtypes (fibroblasts, stromal). In this paper, we propose to use inter-modality learning to label previously un-labelable cell types on virtual H&E. We leveraged multiplexed immunofluorescence (MxIF) histology imaging to identify 14 subclasses of cell types. We performed style transfer to synthesize virtual H&E from MxIF and transferred the higher density labels from MxIF to these virtual H&E images. We then evaluated the efficacy of learning in this approach. We identified helper T and progenitor nuclei with positive predictive values of $0.34 \pm 0.15$ (prevalence $0.03 \pm 0.01$) and $0.47 \pm 0.1$ (prevalence $0.07 \pm 0.02$) respectively on virtual H&E. This approach represents a promising step towards automating annotation in digital pathology.
Deep conditional generative models for longitudinal single-slice abdominal computed tomography harmonization
Sep 17, 2023Abstract:Two-dimensional single-slice abdominal computed tomography (CT) provides a detailed tissue map with high resolution allowing quantitative characterization of relationships between health conditions and aging. However, longitudinal analysis of body composition changes using these scans is difficult due to positional variation between slices acquired in different years, which leading to different organs/tissues captured. To address this issue, we propose C-SliceGen, which takes an arbitrary axial slice in the abdominal region as a condition and generates a pre-defined vertebral level slice by estimating structural changes in the latent space. Our experiments on 2608 volumetric CT data from two in-house datasets and 50 subjects from the 2015 Multi-Atlas Abdomen Labeling Challenge dataset (BTCV) Challenge demonstrate that our model can generate high-quality images that are realistic and similar. We further evaluate our method's capability to harmonize longitudinal positional variation on 1033 subjects from the Baltimore Longitudinal Study of Aging (BLSA) dataset, which contains longitudinal single abdominal slices, and confirmed that our method can harmonize the slice positional variance in terms of visceral fat area. This approach provides a promising direction for mapping slices from different vertebral levels to a target slice and reducing positional variance for single-slice longitudinal analysis. The source code is available at: https://github.com/MASILab/C-SliceGen.
Exploring shared memory architectures for end-to-end gigapixel deep learning
Apr 24, 2023

Abstract:Deep learning has made great strides in medical imaging, enabled by hardware advances in GPUs. One major constraint for the development of new models has been the saturation of GPU memory resources during training. This is especially true in computational pathology, where images regularly contain more than 1 billion pixels. These pathological images are traditionally divided into small patches to enable deep learning due to hardware limitations. In this work, we explore whether the shared GPU/CPU memory architecture on the M1 Ultra systems-on-a-chip (SoCs) recently released by Apple, Inc. may provide a solution. These affordable systems (less than \$5000) provide access to 128 GB of unified memory (Mac Studio with M1 Ultra SoC). As a proof of concept for gigapixel deep learning, we identified tissue from background on gigapixel areas from whole slide images (WSIs). The model was a modified U-Net (4492 parameters) leveraging large kernels and high stride. The M1 Ultra SoC was able to train the model directly on gigapixel images (16000$\times$64000 pixels, 1.024 billion pixels) with a batch size of 1 using over 100 GB of unified memory for the process at an average speed of 1 minute and 21 seconds per batch with Tensorflow 2/Keras. As expected, the model converged with a high Dice score of 0.989 $\pm$ 0.005. Training up until this point took 111 hours and 24 minutes over 4940 steps. Other high RAM GPUs like the NVIDIA A100 (largest commercially accessible at 80 GB, $\sim$\$15000) are not yet widely available (in preview for select regions on Amazon Web Services at \$40.96/hour as a group of 8). This study is a promising step towards WSI-wise end-to-end deep learning with prevalent network architectures.
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Apr 10, 2023



Abstract:The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers.
Scaling Up 3D Kernels with Bayesian Frequency Re-parameterization for Medical Image Segmentation
Mar 10, 2023



Abstract:With the inspiration of vision transformers, the concept of depth-wise convolution revisits to provide a large Effective Receptive Field (ERF) using Large Kernel (LK) sizes for medical image segmentation. However, the segmentation performance might be saturated and even degraded as the kernel sizes scaled up (e.g., $21\times 21\times 21$) in a Convolutional Neural Network (CNN). We hypothesize that convolution with LK sizes is limited to maintain an optimal convergence for locality learning. While Structural Re-parameterization (SR) enhances the local convergence with small kernels in parallel, optimal small kernel branches may hinder the computational efficiency for training. In this work, we propose RepUX-Net, a pure CNN architecture with a simple large kernel block design, which competes favorably with current network state-of-the-art (SOTA) (e.g., 3D UX-Net, SwinUNETR) using 6 challenging public datasets. We derive an equivalency between kernel re-parameterization and the branch-wise variation in kernel convergence. Inspired by the spatial frequency in the human visual system, we extend to vary the kernel convergence into element-wise setting and model the spatial frequency as a Bayesian prior to re-parameterize convolutional weights during training. Specifically, a reciprocal function is leveraged to estimate a frequency-weighted value, which rescales the corresponding kernel element for stochastic gradient descent. From the experimental results, RepUX-Net consistently outperforms 3D SOTA benchmarks with internal validation (FLARE: 0.929 to 0.944), external validation (MSD: 0.901 to 0.932, KiTS: 0.815 to 0.847, LiTS: 0.933 to 0.949, TCIA: 0.736 to 0.779) and transfer learning (AMOS: 0.880 to 0.911) scenarios in Dice Score.
Single Slice Thigh CT Muscle Group Segmentation with Domain Adaptation and Self-Training
Nov 30, 2022Abstract:Objective: Thigh muscle group segmentation is important for assessment of muscle anatomy, metabolic disease and aging. Many efforts have been put into quantifying muscle tissues with magnetic resonance (MR) imaging including manual annotation of individual muscles. However, leveraging publicly available annotations in MR images to achieve muscle group segmentation on single slice computed tomography (CT) thigh images is challenging. Method: We propose an unsupervised domain adaptation pipeline with self-training to transfer labels from 3D MR to single CT slice. First, we transform the image appearance from MR to CT with CycleGAN and feed the synthesized CT images to a segmenter simultaneously. Single CT slices are divided into hard and easy cohorts based on the entropy of pseudo labels inferenced by the segmenter. After refining easy cohort pseudo labels based on anatomical assumption, self-training with easy and hard splits is applied to fine tune the segmenter. Results: On 152 withheld single CT thigh images, the proposed pipeline achieved a mean Dice of 0.888(0.041) across all muscle groups including sartorius, hamstrings, quadriceps femoris and gracilis. muscles Conclusion: To our best knowledge, this is the first pipeline to achieve thigh imaging domain adaptation from MR to CT. The proposed pipeline is effective and robust in extracting muscle groups on 2D single slice CT thigh images.The container is available for public use at https://github.com/MASILab/DA_CT_muscle_seg
Adaptive Contrastive Learning with Dynamic Correlation for Multi-Phase Organ Segmentation
Oct 16, 2022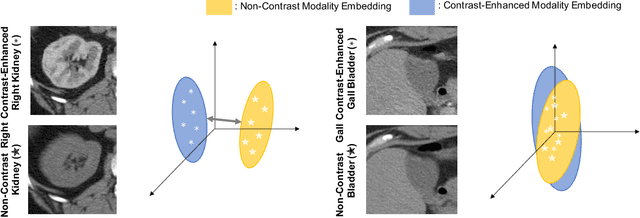
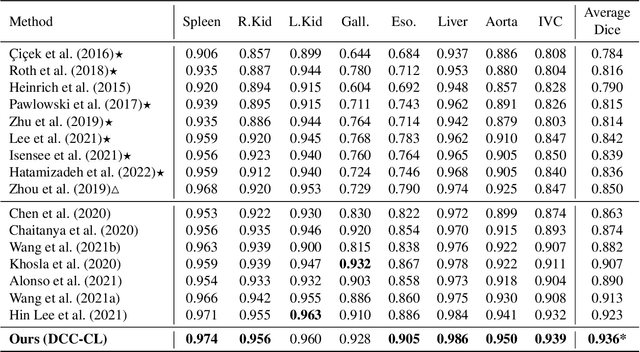
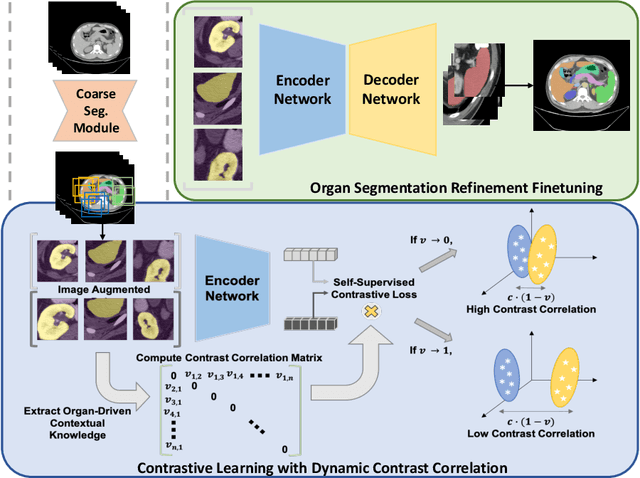
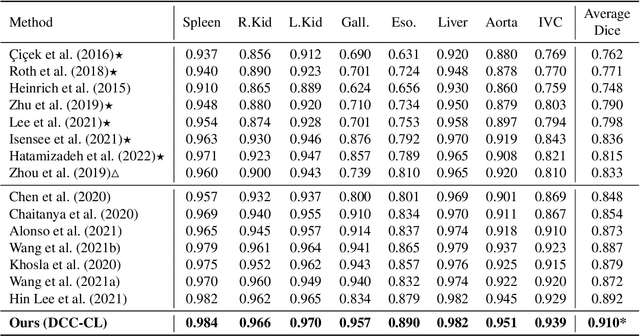
Abstract:Recent studies have demonstrated the superior performance of introducing ``scan-wise" contrast labels into contrastive learning for multi-organ segmentation on multi-phase computed tomography (CT). However, such scan-wise labels are limited: (1) a coarse classification, which could not capture the fine-grained ``organ-wise" contrast variations across all organs; (2) the label (i.e., contrast phase) is typically manually provided, which is error-prone and may introduce manual biases of defining phases. In this paper, we propose a novel data-driven contrastive loss function that adapts the similar/dissimilar contrast relationship between samples in each minibatch at organ-level. Specifically, as variable levels of contrast exist between organs, we hypothesis that the contrast differences in the organ-level can bring additional context for defining representations in the latent space. An organ-wise contrast correlation matrix is computed with mean organ intensities under one-hot attention maps. The goal of adapting the organ-driven correlation matrix is to model variable levels of feature separability at different phases. We evaluate our proposed approach on multi-organ segmentation with both non-contrast CT (NCCT) datasets and the MICCAI 2015 BTCV Challenge contrast-enhance CT (CECT) datasets. Compared to the state-of-the-art approaches, our proposed contrastive loss yields a substantial and significant improvement of 1.41% (from 0.923 to 0.936, p-value$<$0.01) and 2.02% (from 0.891 to 0.910, p-value$<$0.01) on mean Dice scores across all organs with respect to NCCT and CECT cohorts. We further assess the trained model performance with the MICCAI 2021 FLARE Challenge CECT datasets and achieve a substantial improvement of mean Dice score from 0.927 to 0.934 (p-value$<$0.01). The code is available at: https://github.com/MASILab/DCC_CL
UNesT: Local Spatial Representation Learning with Hierarchical Transformer for Efficient Medical Segmentation
Sep 28, 2022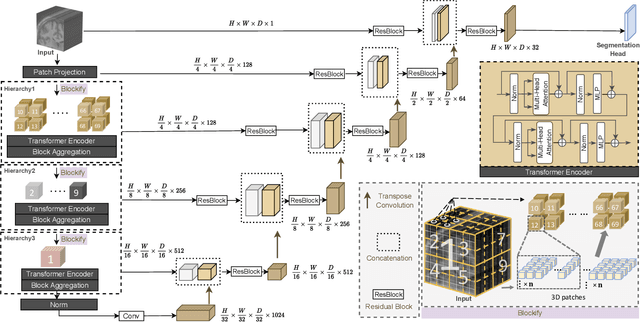
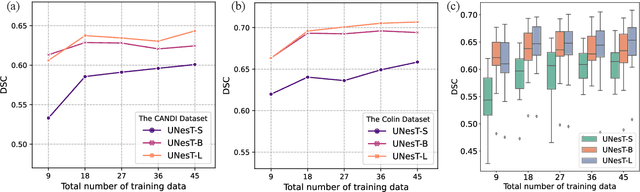
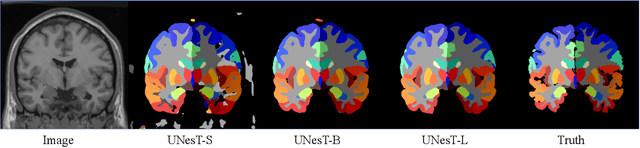
Abstract:Transformer-based models, capable of learning better global dependencies, have recently demonstrated exceptional representation learning capabilities in computer vision and medical image analysis. Transformer reformats the image into separate patches and realize global communication via the self-attention mechanism. However, positional information between patches is hard to preserve in such 1D sequences, and loss of it can lead to sub-optimal performance when dealing with large amounts of heterogeneous tissues of various sizes in 3D medical image segmentation. Additionally, current methods are not robust and efficient for heavy-duty medical segmentation tasks such as predicting a large number of tissue classes or modeling globally inter-connected tissues structures. Inspired by the nested hierarchical structures in vision transformer, we proposed a novel 3D medical image segmentation method (UNesT), employing a simplified and faster-converging transformer encoder design that achieves local communication among spatially adjacent patch sequences by aggregating them hierarchically. We extensively validate our method on multiple challenging datasets, consisting anatomies of 133 structures in brain, 14 organs in abdomen, 4 hierarchical components in kidney, and inter-connected kidney tumors). We show that UNesT consistently achieves state-of-the-art performance and evaluate its generalizability and data efficiency. Particularly, the model achieves whole brain segmentation task complete ROI with 133 tissue classes in single network, outperforms prior state-of-the-art method SLANT27 ensembled with 27 network tiles, our model performance increases the mean DSC score of the publicly available Colin and CANDI dataset from 0.7264 to 0.7444 and from 0.6968 to 0.7025, respectively.
Joint analysis of structural connectivity and cortical surface features: correlates with mild traumatic brain injury
Dec 15, 2020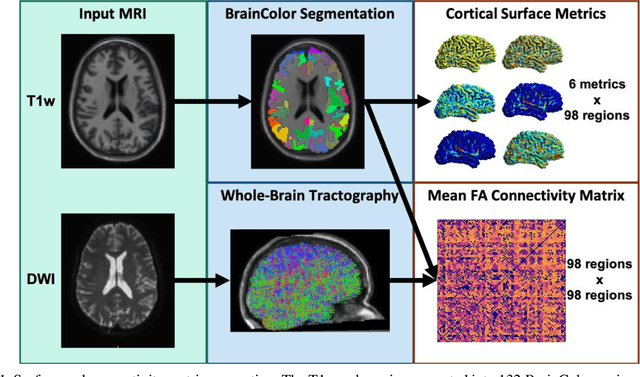
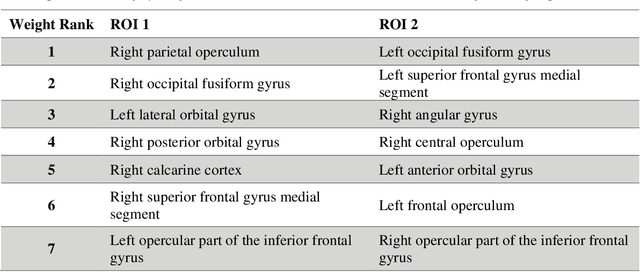
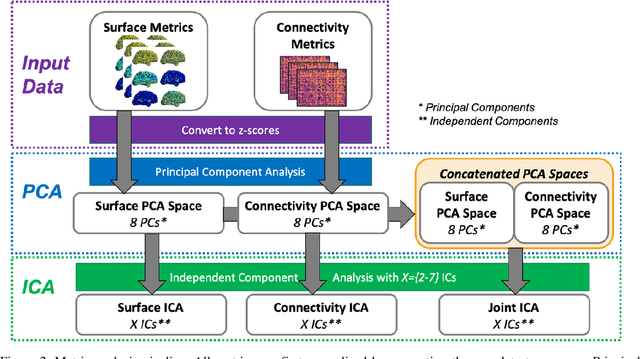
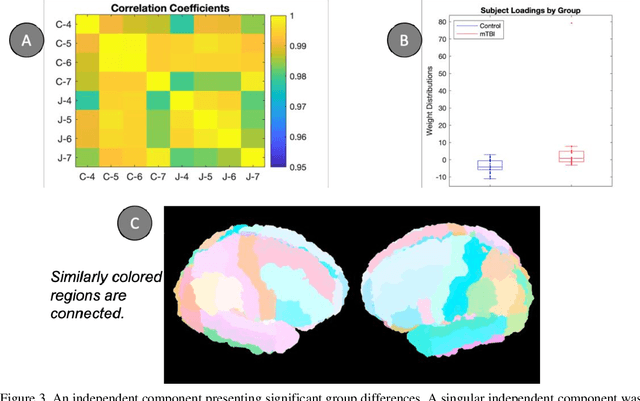
Abstract:Mild traumatic brain injury (mTBI) is a complex syndrome that affects up to 600 per 100,000 individuals, with a particular concentration among military personnel. About half of all mTBI patients experience a diverse array of chronic symptoms which persist long after the acute injury. Hence, there is an urgent need for better understanding of the white matter and gray matter pathologies associated with mTBI to map which specific brain systems are impacted and identify courses of intervention. Previous works have linked mTBI to disruptions in white matter pathways and cortical surface abnormalities. Herein, we examine these hypothesized links in an exploratory study of joint structural connectivity and cortical surface changes associated with mTBI and its chronic symptoms. Briefly, we consider a cohort of 12 mTBI and 26 control subjects. A set of 588 cortical surface metrics and 4,753 structural connectivity metrics were extracted from cortical surface regions and diffusion weighted magnetic resonance imaging in each subject. Principal component analysis (PCA) was used to reduce the dimensionality of each metric set. We then applied independent component analysis (ICA) both to each PCA space individually and together in a joint ICA approach. We identified a stable independent component across the connectivity-only and joint ICAs which presented significant group differences in subject loadings (p<0.05, corrected). Additionally, we found that two mTBI symptoms, slowed thinking and forgetfulness, were significantly correlated (p<0.05, corrected) with mTBI subject loadings in a surface-only ICA. These surface-only loadings captured an increase in bilateral cortical thickness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge