Karol Gotkowski
Benchmark of Segmentation Techniques for Pelvic Fracture in CT and X-ray: Summary of the PENGWIN 2024 Challenge
Apr 03, 2025Abstract:The segmentation of pelvic fracture fragments in CT and X-ray images is crucial for trauma diagnosis, surgical planning, and intraoperative guidance. However, accurately and efficiently delineating the bone fragments remains a significant challenge due to complex anatomy and imaging limitations. The PENGWIN challenge, organized as a MICCAI 2024 satellite event, aimed to advance automated fracture segmentation by benchmarking state-of-the-art algorithms on these complex tasks. A diverse dataset of 150 CT scans was collected from multiple clinical centers, and a large set of simulated X-ray images was generated using the DeepDRR method. Final submissions from 16 teams worldwide were evaluated under a rigorous multi-metric testing scheme. The top-performing CT algorithm achieved an average fragment-wise intersection over union (IoU) of 0.930, demonstrating satisfactory accuracy. However, in the X-ray task, the best algorithm attained an IoU of 0.774, highlighting the greater challenges posed by overlapping anatomical structures. Beyond the quantitative evaluation, the challenge revealed methodological diversity in algorithm design. Variations in instance representation, such as primary-secondary classification versus boundary-core separation, led to differing segmentation strategies. Despite promising results, the challenge also exposed inherent uncertainties in fragment definition, particularly in cases of incomplete fractures. These findings suggest that interactive segmentation approaches, integrating human decision-making with task-relevant information, may be essential for improving model reliability and clinical applicability.
Embarrassingly Simple Scribble Supervision for 3D Medical Segmentation
Mar 19, 2024



Abstract:Traditionally, segmentation algorithms require dense annotations for training, demanding significant annotation efforts, particularly within the 3D medical imaging field. Scribble-supervised learning emerges as a possible solution to this challenge, promising a reduction in annotation efforts when creating large-scale datasets. Recently, a plethora of methods for optimized learning from scribbles have been proposed, but have so far failed to position scribble annotation as a beneficial alternative. We relate this shortcoming to two major issues: 1) the complex nature of many methods which deeply ties them to the underlying segmentation model, thus preventing a migration to more powerful state-of-the-art models as the field progresses and 2) the lack of a systematic evaluation to validate consistent performance across the broader medical domain, resulting in a lack of trust when applying these methods to new segmentation problems. To address these issues, we propose a comprehensive scribble supervision benchmark consisting of seven datasets covering a diverse set of anatomies and pathologies imaged with varying modalities. We furthermore propose the systematic use of partial losses, i.e. losses that are only computed on annotated voxels. Contrary to most existing methods, these losses can be seamlessly integrated into state-of-the-art segmentation methods, enabling them to learn from scribble annotations while preserving their original loss formulations. Our evaluation using nnU-Net reveals that while most existing methods suffer from a lack of generalization, the proposed approach consistently delivers state-of-the-art performance. Thanks to its simplicity, our approach presents an embarrassingly simple yet effective solution to the challenges of scribble supervision. Source code as well as our extensive scribble benchmarking suite will be made publicly available upon publication.
Scalable, out-of-the box segmentation of individual particles from mineral samples acquired with micro CT
Feb 13, 2023Abstract:Minerals are indispensable for a functioning modern society. Yet, their supply is limited causing a need for optimizing their exploration and extraction both from ores and recyclable materials. Typically, these processes must be meticulously adapted to the precise properties of the processed particles, requiring an extensive characterization of their shapes, appearances as well as the overall material composition. Current approaches perform this analysis based on bulk segmentation and characterization of particles, and rely on rudimentary postprocessing techniques to separate touching particles. However, due to their inability to reliably perform this separation as well as the need to retrain or reconfigure most methods for each new image, these approaches leave untapped potential to be leveraged. Here, we propose an instance segmentation method that is able to extract individual particles from large micro CT images taken from mineral samples embedded in an epoxy matrix. Our approach is based on the powerful nnU-Net framework, introduces a particle size normalization, makes use of a border-core representation to enable instance segmentation and is trained with a large dataset containing particles of numerous different materials and minerals. We demonstrate that our approach can be applied out-of-the box to a large variety of particle types, including materials and appearances that have not been part of the training set. Thus, no further manual annotations and retraining are required when applying the method to new mineral samples, enabling substantially higher scalability of experiments than existing methods. Our code and dataset are made publicly available.
Distance-based detection of out-of-distribution silent failures for Covid-19 lung lesion segmentation
Aug 05, 2022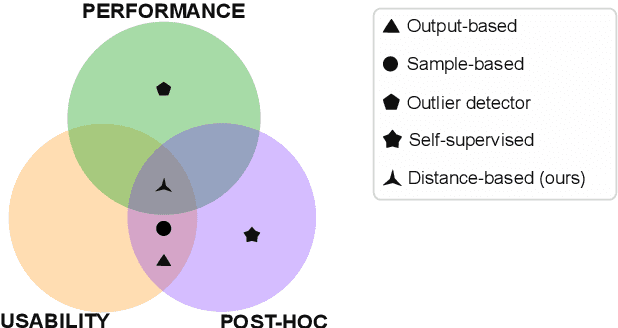
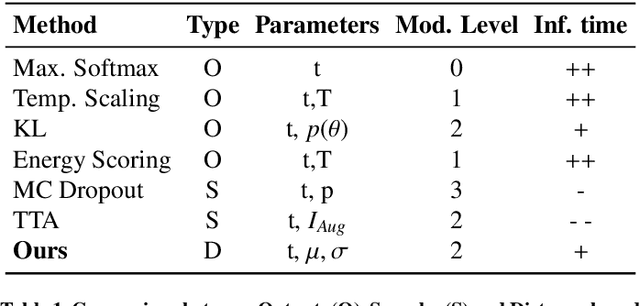
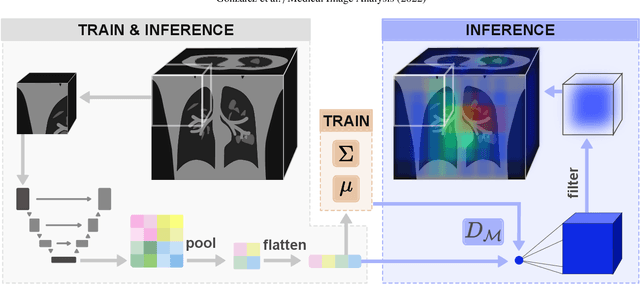
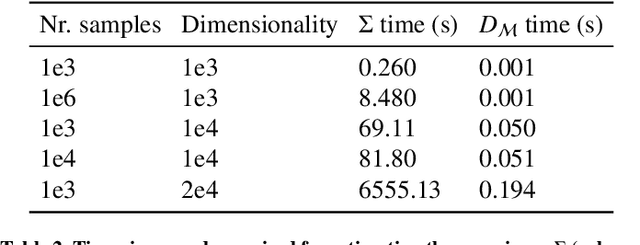
Abstract:Automatic segmentation of ground glass opacities and consolidations in chest computer tomography (CT) scans can potentially ease the burden of radiologists during times of high resource utilisation. However, deep learning models are not trusted in the clinical routine due to failing silently on out-of-distribution (OOD) data. We propose a lightweight OOD detection method that leverages the Mahalanobis distance in the feature space and seamlessly integrates into state-of-the-art segmentation pipelines. The simple approach can even augment pre-trained models with clinically relevant uncertainty quantification. We validate our method across four chest CT distribution shifts and two magnetic resonance imaging applications, namely segmentation of the hippocampus and the prostate. Our results show that the proposed method effectively detects far- and near-OOD samples across all explored scenarios.
Detecting when pre-trained nnU-Net models fail silently for Covid-19 lung lesion segmentation
Jul 14, 2021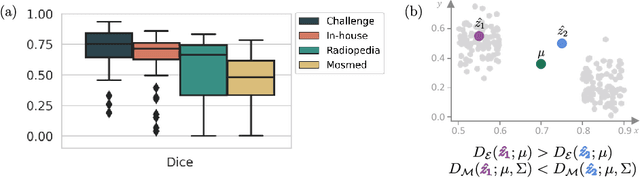
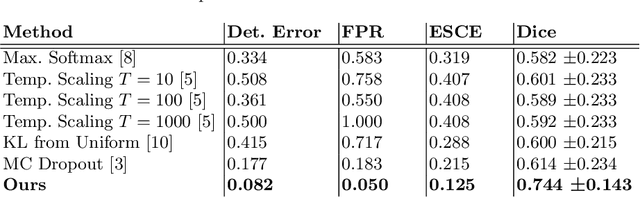
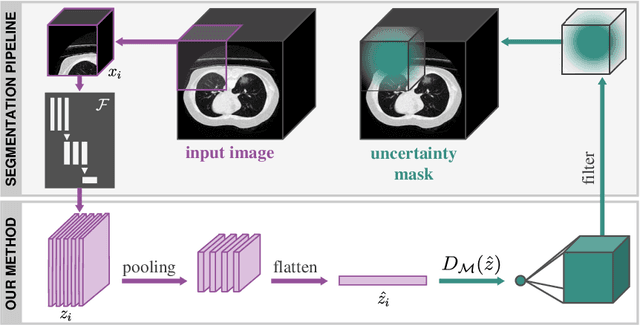
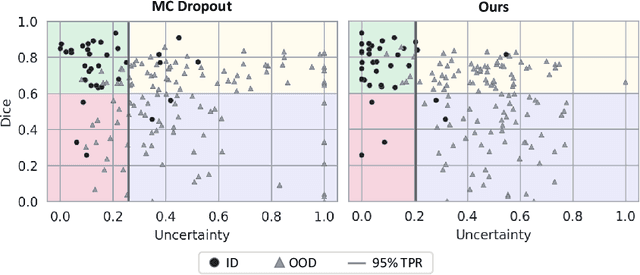
Abstract:Automatic segmentation of lung lesions in computer tomography has the potential to ease the burden of clinicians during the Covid-19 pandemic. Yet predictive deep learning models are not trusted in the clinical routine due to failing silently in out-of-distribution (OOD) data. We propose a lightweight OOD detection method that exploits the Mahalanobis distance in the feature space. The proposed approach can be seamlessly integrated into state-of-the-art segmentation pipelines without requiring changes in model architecture or training procedure, and can therefore be used to assess the suitability of pre-trained models to new data. We validate our method with a patch-based nnU-Net architecture trained with a multi-institutional dataset and find that it effectively detects samples that the model segments incorrectly.
M3d-CAM: A PyTorch library to generate 3D data attention maps for medical deep learning
Jul 01, 2020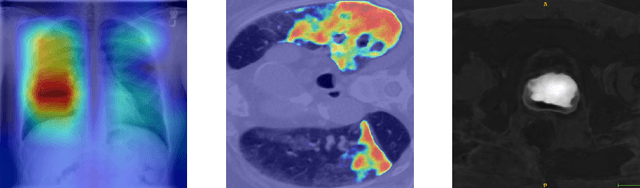
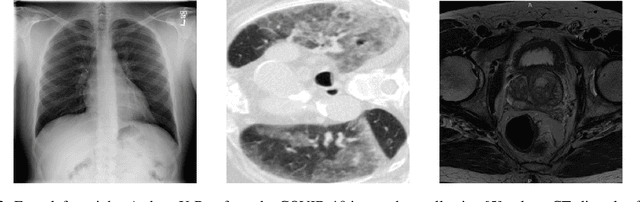
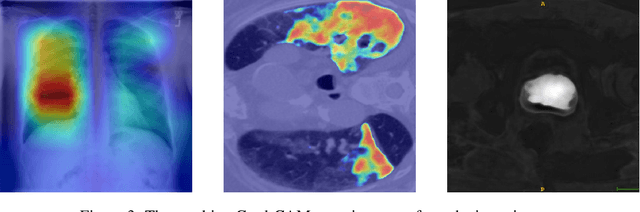
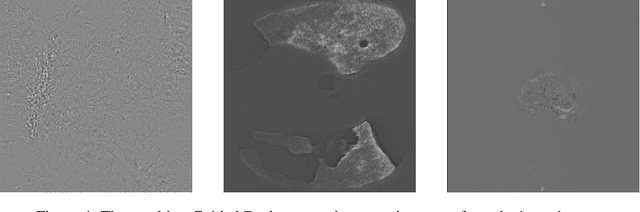
Abstract:M3d-CAM is an easy to use library for generating attention maps of CNN-based PyTorch models improving the interpretability of model predictions for humans. The attention maps can be generated with multiple methods like Guided Backpropagation, Grad-CAM, Guided Grad-CAM and Grad-CAM++. These attention maps visualize the regions in the input data that influenced the model prediction the most at a certain layer. Furthermore, M3d-CAM supports 2D and 3D data for the task of classification as well as for segmentation. A key feature is also that in most cases only a single line of code is required for generating attention maps for a model making M3d-CAM basically plug and play.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge