Jianpeng Zhang
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023



Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
Attention Mechanisms in Medical Image Segmentation: A Survey
May 29, 2023Abstract:Medical image segmentation plays an important role in computer-aided diagnosis. Attention mechanisms that distinguish important parts from irrelevant parts have been widely used in medical image segmentation tasks. This paper systematically reviews the basic principles of attention mechanisms and their applications in medical image segmentation. First, we review the basic concepts of attention mechanism and formulation. Second, we surveyed over 300 articles related to medical image segmentation, and divided them into two groups based on their attention mechanisms, non-Transformer attention and Transformer attention. In each group, we deeply analyze the attention mechanisms from three aspects based on the current literature work, i.e., the principle of the mechanism (what to use), implementation methods (how to use), and application tasks (where to use). We also thoroughly analyzed the advantages and limitations of their applications to different tasks. Finally, we summarize the current state of research and shortcomings in the field, and discuss the potential challenges in the future, including task specificity, robustness, standard evaluation, etc. We hope that this review can showcase the overall research context of traditional and Transformer attention methods, provide a clear reference for subsequent research, and inspire more advanced attention research, not only in medical image segmentation, but also in other image analysis scenarios.
UniSeg: A Prompt-driven Universal Segmentation Model as well as A Strong Representation Learner
Apr 07, 2023Abstract:The universal model emerges as a promising trend for medical image segmentation, paving up the way to build medical imaging large model (MILM). One popular strategy to build universal models is to encode each task as a one-hot vector and generate dynamic convolutional layers at the end of the decoder to extract the interested target. Although successful, it ignores the correlations among tasks and meanwhile is too late to make the model 'aware' of the ongoing task. To address both issues, we propose a prompt-driven Universal Segmentation model (UniSeg) for multi-task medical image segmentation using diverse modalities and domains. We first devise a learnable universal prompt to describe the correlations among all tasks and then convert this prompt and image features into a task-specific prompt, which is fed to the decoder as a part of its input. Thus, we make the model 'aware' of the ongoing task early and boost the task-specific training of the whole decoder. Our results indicate that the proposed UniSeg outperforms other universal models and single-task models on 11 upstream tasks. Moreover, UniSeg also beats other pre-trained models on two downstream datasets, providing the community with a high-quality pre-trained model for 3D medical image segmentation. Code and model are available at https://github.com/yeerwen/UniSeg.
Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans
Jan 28, 2023



Abstract:Human readers or radiologists routinely perform full-body multi-organ multi-disease detection and diagnosis in clinical practice, while most medical AI systems are built to focus on single organs with a narrow list of a few diseases. This might severely limit AI's clinical adoption. A certain number of AI models need to be assembled non-trivially to match the diagnostic process of a human reading a CT scan. In this paper, we construct a Unified Tumor Transformer (UniT) model to detect (tumor existence and location) and diagnose (tumor characteristics) eight major cancer-prevalent organs in CT scans. UniT is a query-based Mask Transformer model with the output of multi-organ and multi-tumor semantic segmentation. We decouple the object queries into organ queries, detection queries and diagnosis queries, and further establish hierarchical relationships among the three groups. This clinically-inspired architecture effectively assists inter- and intra-organ representation learning of tumors and facilitates the resolution of these complex, anatomically related multi-organ cancer image reading tasks. UniT is trained end-to-end using a curated large-scale CT images of 10,042 patients including eight major types of cancers and occurring non-cancer tumors (all are pathology-confirmed with 3D tumor masks annotated by radiologists). On the test set of 631 patients, UniT has demonstrated strong performance under a set of clinically relevant evaluation metrics, substantially outperforming both multi-organ segmentation methods and an assembly of eight single-organ expert models in tumor detection, segmentation, and diagnosis. Such a unified multi-cancer image reading model (UniT) can significantly reduce the number of false positives produced by combined multi-system models. This moves one step closer towards a universal high-performance cancer screening tool.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Learning from partially labeled data for multi-organ and tumor segmentation
Nov 13, 2022Abstract:Medical image benchmarks for the segmentation of organs and tumors suffer from the partially labeling issue due to its intensive cost of labor and expertise. Current mainstream approaches follow the practice of one network solving one task. With this pipeline, not only the performance is limited by the typically small dataset of a single task, but also the computation cost linearly increases with the number of tasks. To address this, we propose a Transformer based dynamic on-demand network (TransDoDNet) that learns to segment organs and tumors on multiple partially labeled datasets. Specifically, TransDoDNet has a hybrid backbone that is composed of the convolutional neural network and Transformer. A dynamic head enables the network to accomplish multiple segmentation tasks flexibly. Unlike existing approaches that fix kernels after training, the kernels in the dynamic head are generated adaptively by the Transformer, which employs the self-attention mechanism to model long-range organ-wise dependencies and decodes the organ embedding that can represent each organ. We create a large-scale partially labeled Multi-Organ and Tumor Segmentation benchmark, termed MOTS, and demonstrate the superior performance of our TransDoDNet over other competitors on seven organ and tumor segmentation tasks. This study also provides a general 3D medical image segmentation model, which has been pre-trained on the large-scale MOTS benchmark and has demonstrated advanced performance over BYOL, the current predominant self-supervised learning method. Code will be available at \url{https://git.io/DoDNet}.
ClusTR: Exploring Efficient Self-attention via Clustering for Vision Transformers
Aug 28, 2022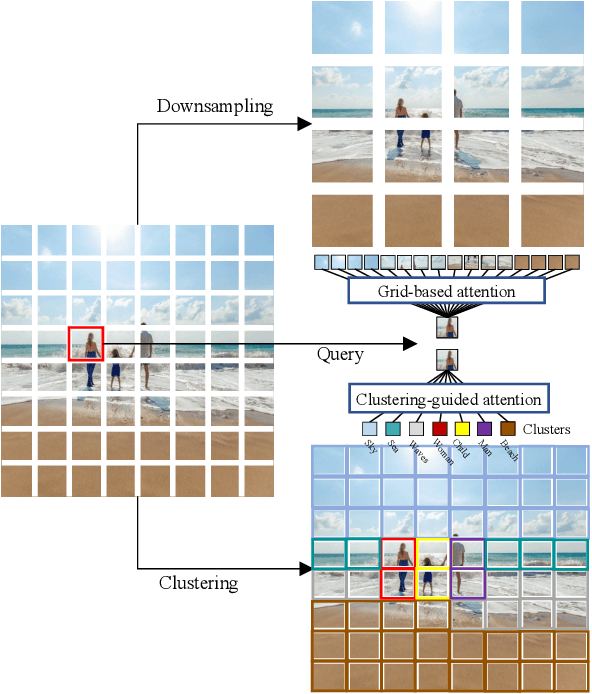
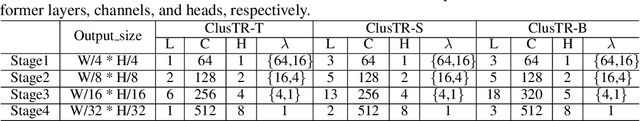
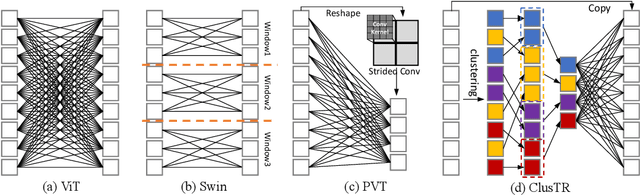

Abstract:Although Transformers have successfully transitioned from their language modelling origins to image-based applications, their quadratic computational complexity remains a challenge, particularly for dense prediction. In this paper we propose a content-based sparse attention method, as an alternative to dense self-attention, aiming to reduce the computation complexity while retaining the ability to model long-range dependencies. Specifically, we cluster and then aggregate key and value tokens, as a content-based method of reducing the total token count. The resulting clustered-token sequence retains the semantic diversity of the original signal, but can be processed at a lower computational cost. Besides, we further extend the clustering-guided attention from single-scale to multi-scale, which is conducive to dense prediction tasks. We label the proposed Transformer architecture ClusTR, and demonstrate that it achieves state-of-the-art performance on various vision tasks but at lower computational cost and with fewer parameters. For instance, our ClusTR small model with 22.7M parameters achieves 83.2\% Top-1 accuracy on ImageNet. Source code and ImageNet models will be made publicly available.
Uncertainty-aware Multi-modal Learning via Cross-modal Random Network Prediction
Jul 22, 2022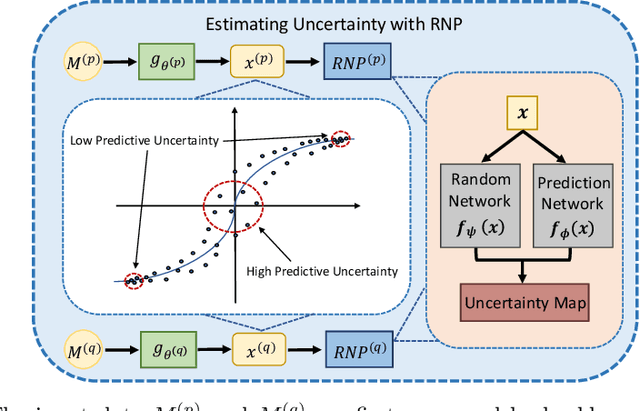
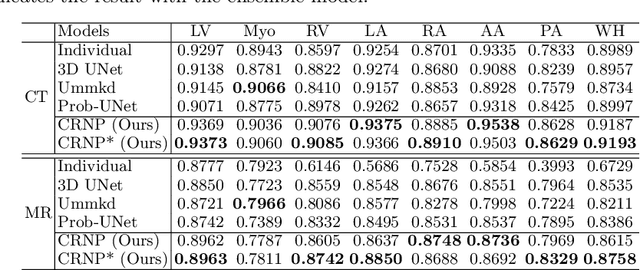
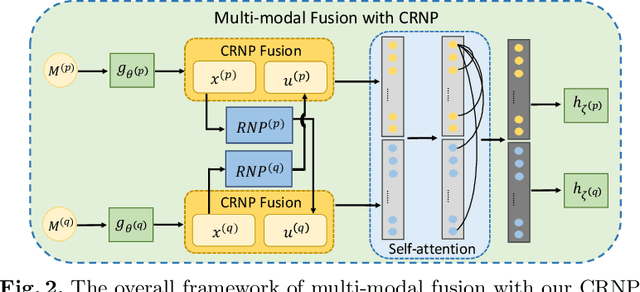
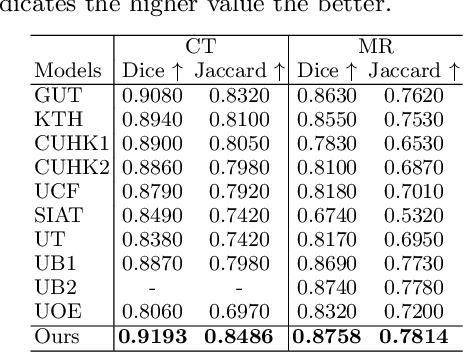
Abstract:Multi-modal learning focuses on training models by equally combining multiple input data modalities during the prediction process. However, this equal combination can be detrimental to the prediction accuracy because different modalities are usually accompanied by varying levels of uncertainty. Using such uncertainty to combine modalities has been studied by a couple of approaches, but with limited success because these approaches are either designed to deal with specific classification or segmentation problems and cannot be easily translated into other tasks, or suffer from numerical instabilities. In this paper, we propose a new Uncertainty-aware Multi-modal Learner that estimates uncertainty by measuring feature density via Cross-modal Random Network Prediction (CRNP). CRNP is designed to require little adaptation to translate between different prediction tasks, while having a stable training process. From a technical point of view, CRNP is the first approach to explore random network prediction to estimate uncertainty and to combine multi-modal data. Experiments on two 3D multi-modal medical image segmentation tasks and three 2D multi-modal computer vision classification tasks show the effectiveness, adaptability and robustness of CRNP. Also, we provide an extensive discussion on different fusion functions and visualization to validate the proposed model.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022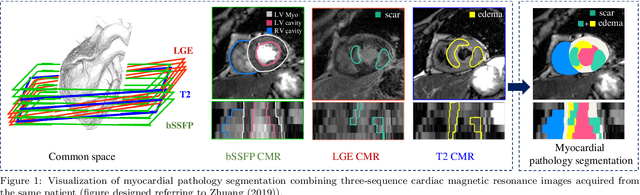
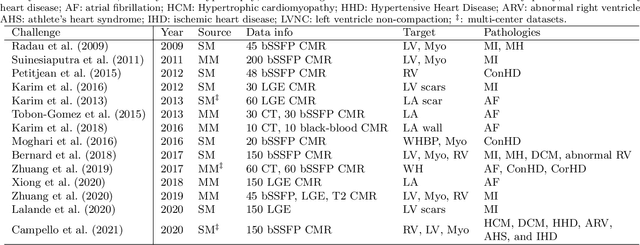
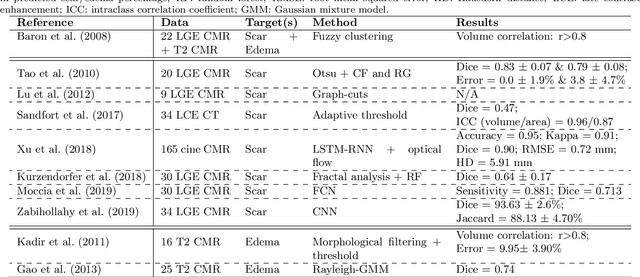
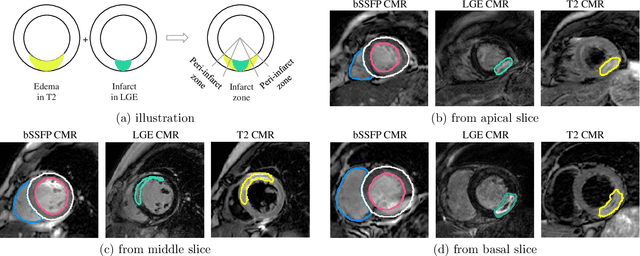
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
Unified 2D and 3D Pre-training for Medical Image classification and Segmentation
Dec 17, 2021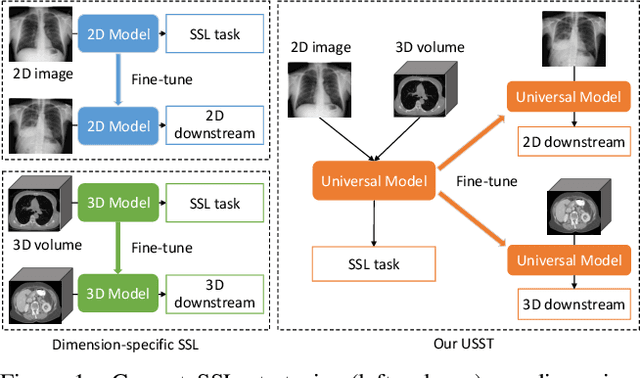
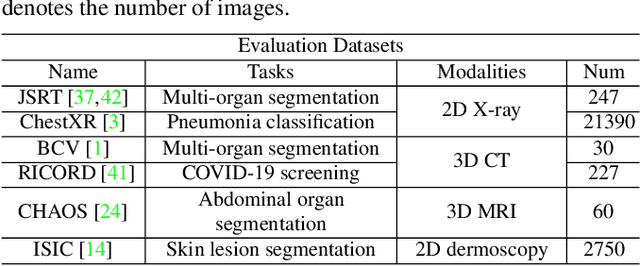
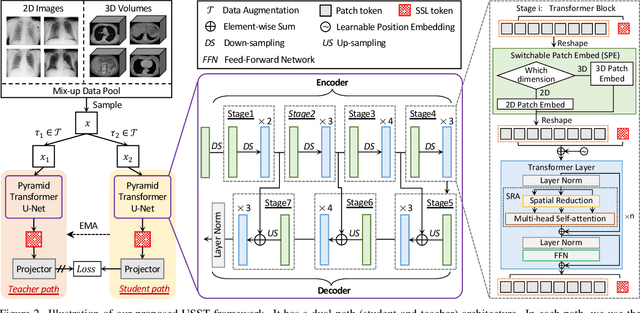
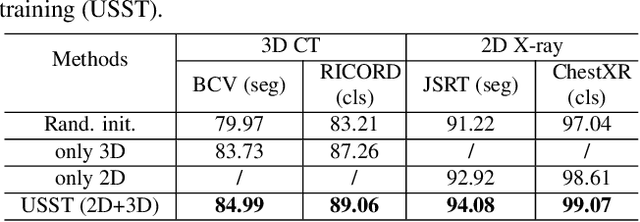
Abstract:Self-supervised learning (SSL) opens up huge opportunities for better utilizing unlabeled data. It is essential for medical image analysis that is generally known for its lack of annotations. However, when we attempt to use as many as possible unlabeled medical images in SSL, breaking the dimension barrier (\ie, making it possible to jointly use both 2D and 3D images) becomes a must. In this paper, we propose a Universal Self-Supervised Transformer (USST) framework based on the student-teacher paradigm, aiming to leverage a huge of unlabeled medical data with multiple dimensions to learn rich representations. To achieve this, we design a Pyramid Transformer U-Net (PTU) as the backbone, which is composed of switchable patch embedding (SPE) layers and Transformer layers. The SPE layer switches to either 2D or 3D patch embedding depending on the input dimension. After that, the images are converted to a sequence regardless of their original dimensions. The Transformer layer then models the long-term dependencies in a sequence-to-sequence manner, thus enabling USST to learn representations from both 2D and 3D images. USST has two obvious merits compared to current dimension-specific SSL: (1) \textbf{more effective} - can learn representations from more and diverse data; and (2) \textbf{more versatile} - can be transferred to various downstream tasks. The results show that USST provides promising results on six 2D/3D medical image classification and segmentation tasks, outperforming the supervised ImageNet pre-training and advanced SSL counterparts substantially.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge