Jian Zhuang
Myocardial Segmentation of Cardiac MRI Sequences with Temporal Consistency for Coronary Artery Disease Diagnosis
Dec 29, 2020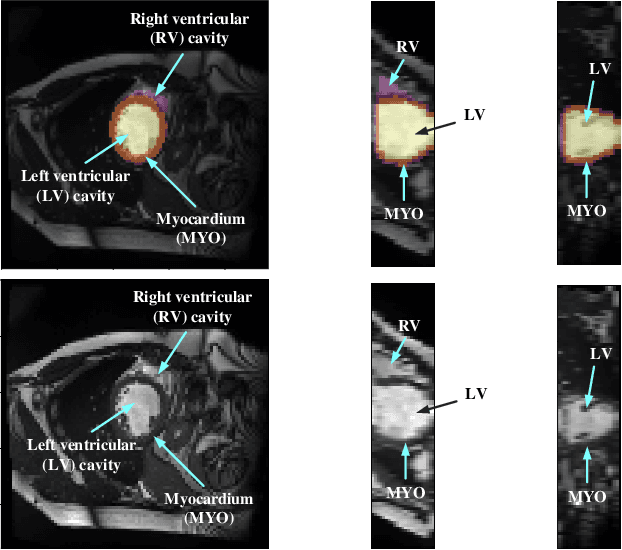
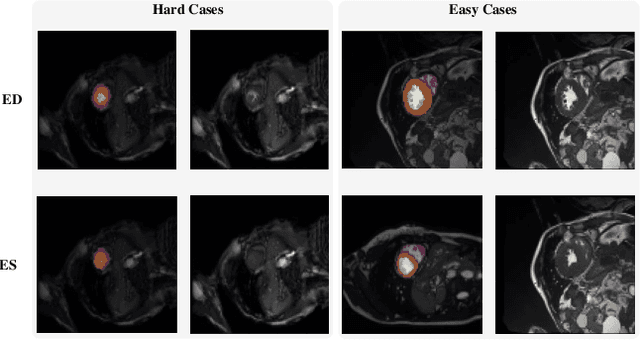
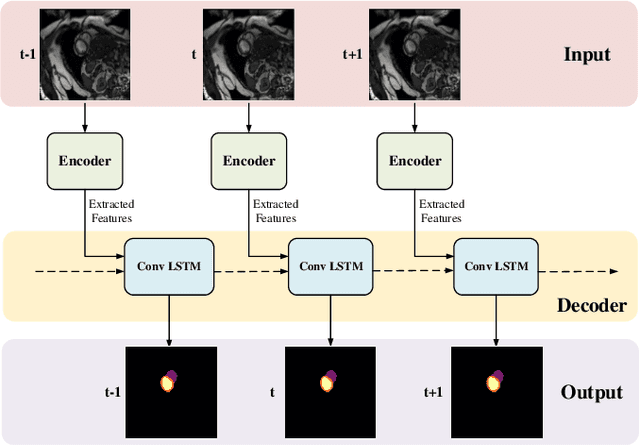
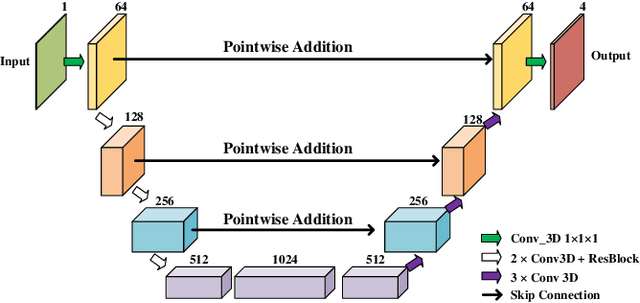
Abstract:Coronary artery disease (CAD) is the most common cause of death globally, and its diagnosis is usually based on manual myocardial segmentation of Magnetic Resonance Imaging (MRI) sequences. As the manual segmentation is tedious, time-consuming and with low applicability, automatic myocardial segmentation using machine learning techniques has been widely explored recently. However, almost all the existing methods treat the input MRI sequences independently, which fails to capture the temporal information between sequences, e.g., the shape and location information of the myocardium in sequences along time. In this paper, we propose a myocardial segmentation framework for sequence of cardiac MRI (CMR) scanning images of left ventricular cavity, right ventricular cavity, and myocardium. Specifically, we propose to combine conventional networks and recurrent networks to incorporate temporal information between sequences to ensure temporal consistent. We evaluated our framework on the Automated Cardiac Diagnosis Challenge (ACDC) dataset. Experiment results demonstrate that our framework can improve the segmentation accuracy by up to 2% in Dice coefficient.
Do Noises Bother Human and Neural Networks In the Same Way? A Medical Image Analysis Perspective
Nov 04, 2020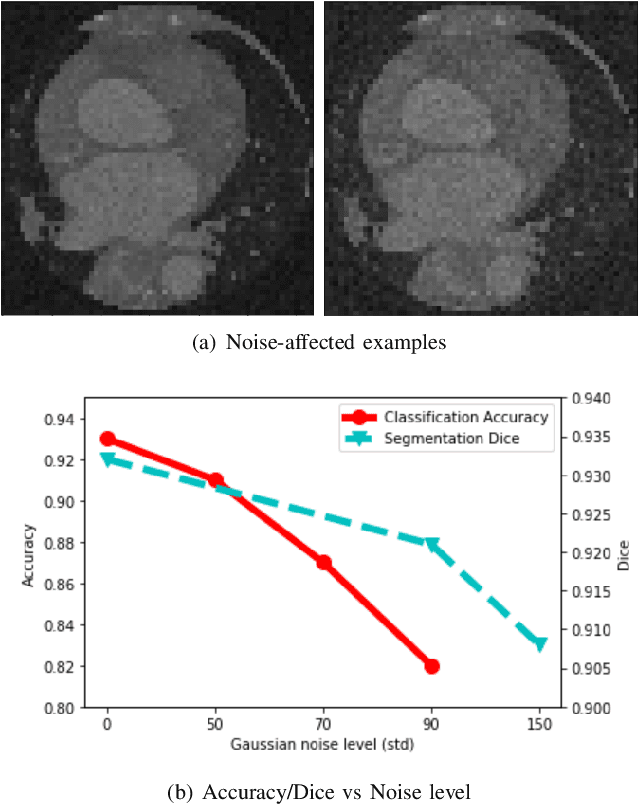
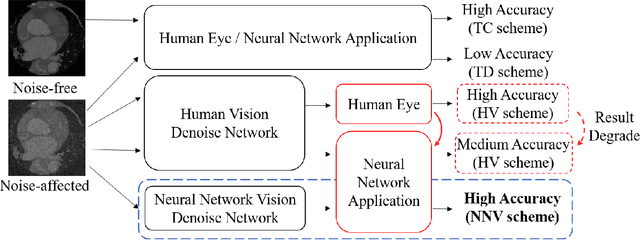
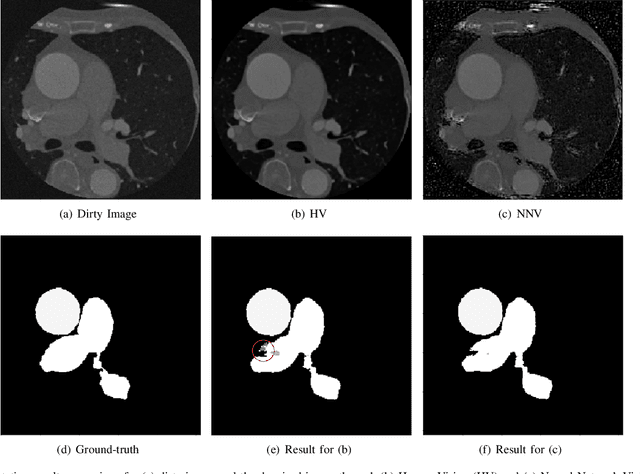
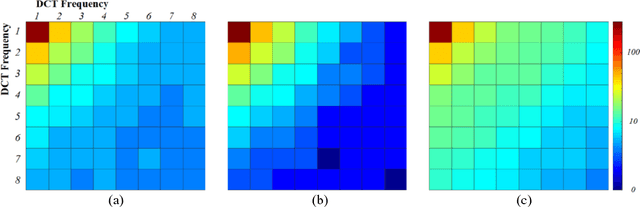
Abstract:Deep learning had already demonstrated its power in medical images, including denoising, classification, segmentation, etc. All these applications are proposed to automatically analyze medical images beforehand, which brings more information to radiologists during clinical assessment for accuracy improvement. Recently, many medical denoising methods had shown their significant artifact reduction result and noise removal both quantitatively and qualitatively. However, those existing methods are developed around human-vision, i.e., they are designed to minimize the noise effect that can be perceived by human eyes. In this paper, we introduce an application-guided denoising framework, which focuses on denoising for the following neural networks. In our experiments, we apply the proposed framework to different datasets, models, and use cases. Experimental results show that our proposed framework can achieve a better result than human-vision denoising network.
Towards Cardiac Intervention Assistance: Hardware-aware Neural Architecture Exploration for Real-Time 3D Cardiac Cine MRI Segmentation
Aug 17, 2020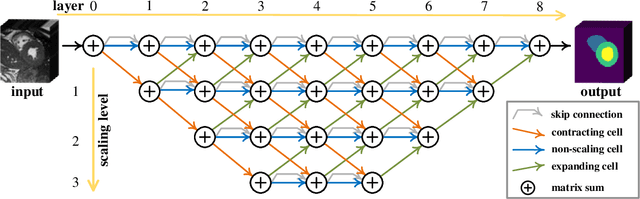
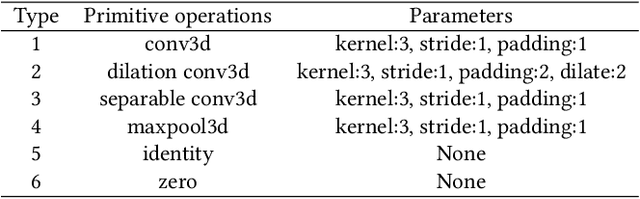
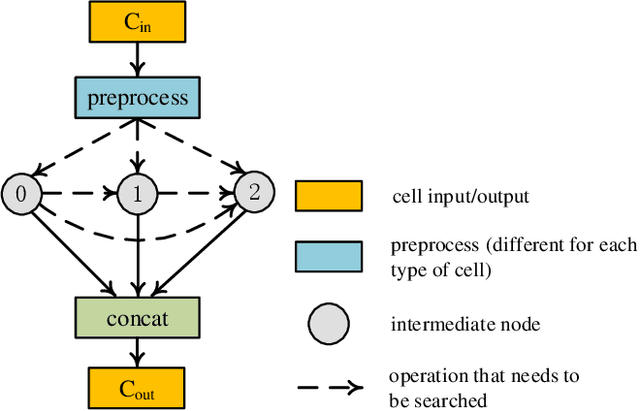
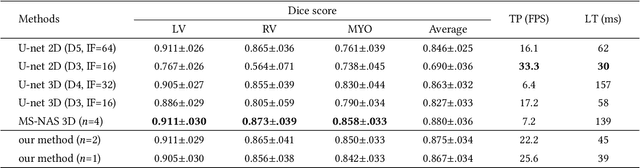
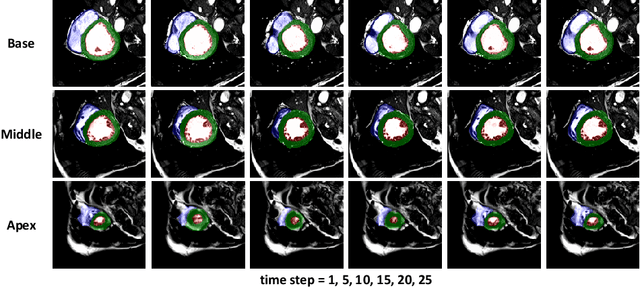
Abstract:Real-time cardiac magnetic resonance imaging (MRI) plays an increasingly important role in guiding various cardiac interventions. In order to provide better visual assistance, the cine MRI frames need to be segmented on-the-fly to avoid noticeable visual lag. In addition, considering reliability and patient data privacy, the computation is preferably done on local hardware. State-of-the-art MRI segmentation methods mostly focus on accuracy only, and can hardly be adopted for real-time application or on local hardware. In this work, we present the first hardware-aware multi-scale neural architecture search (NAS) framework for real-time 3D cardiac cine MRI segmentation. The proposed framework incorporates a latency regularization term into the loss function to handle real-time constraints, with the consideration of underlying hardware. In addition, the formulation is fully differentiable with respect to the architecture parameters, so that stochastic gradient descent (SGD) can be used for optimization to reduce the computation cost while maintaining optimization quality. Experimental results on ACDC MICCAI 2017 dataset demonstrate that our hardware-aware multi-scale NAS framework can reduce the latency by up to 3.5 times and satisfy the real-time constraints, while still achieving competitive segmentation accuracy, compared with the state-of-the-art NAS segmentation framework.
ICA-UNet: ICA Inspired Statistical UNet for Real-time 3D Cardiac Cine MRI Segmentation
Jul 18, 2020
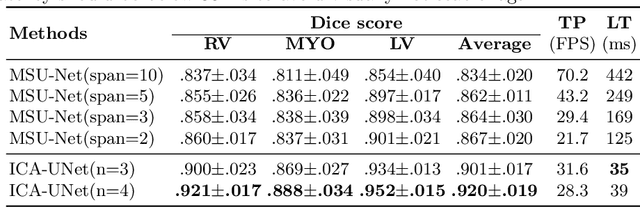
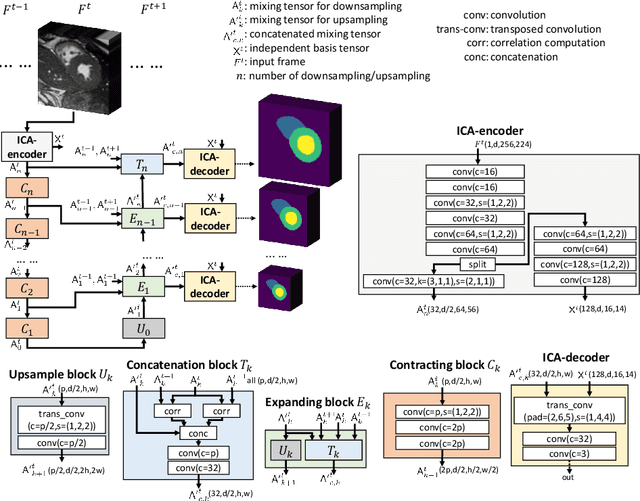
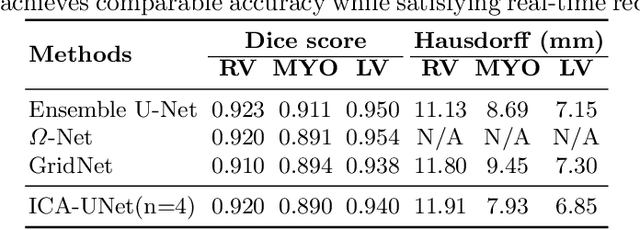
Abstract:Real-time cine magnetic resonance imaging (MRI) plays an increasingly important role in various cardiac interventions. In order to enable fast and accurate visual assistance, the temporal frames need to be segmented on-the-fly. However, state-of-the-art MRI segmentation methods are used either offline because of their high computation complexity, or in real-time but with significant accuracy loss and latency increase (causing visually noticeable lag). As such, they can hardly be adopted to assist visual guidance. In this work, inspired by a new interpretation of Independent Component Analysis (ICA) for learning, we propose a novel ICA-UNet for real-time 3D cardiac cine MRI segmentation. Experiments using the MICCAI ACDC 2017 dataset show that, compared with the state-of-the-arts, ICA-UNet not only achieves higher Dice scores, but also meets the real-time requirements for both throughput and latency (up to 12.6X reduction), enabling real-time guidance for cardiac interventions without visual lag.
Multi-Cycle-Consistent Adversarial Networks for CT Image Denoising
Feb 27, 2020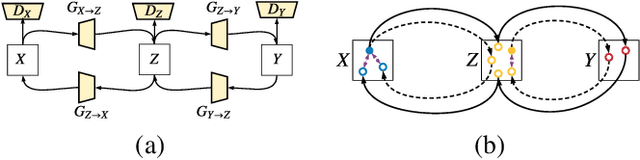
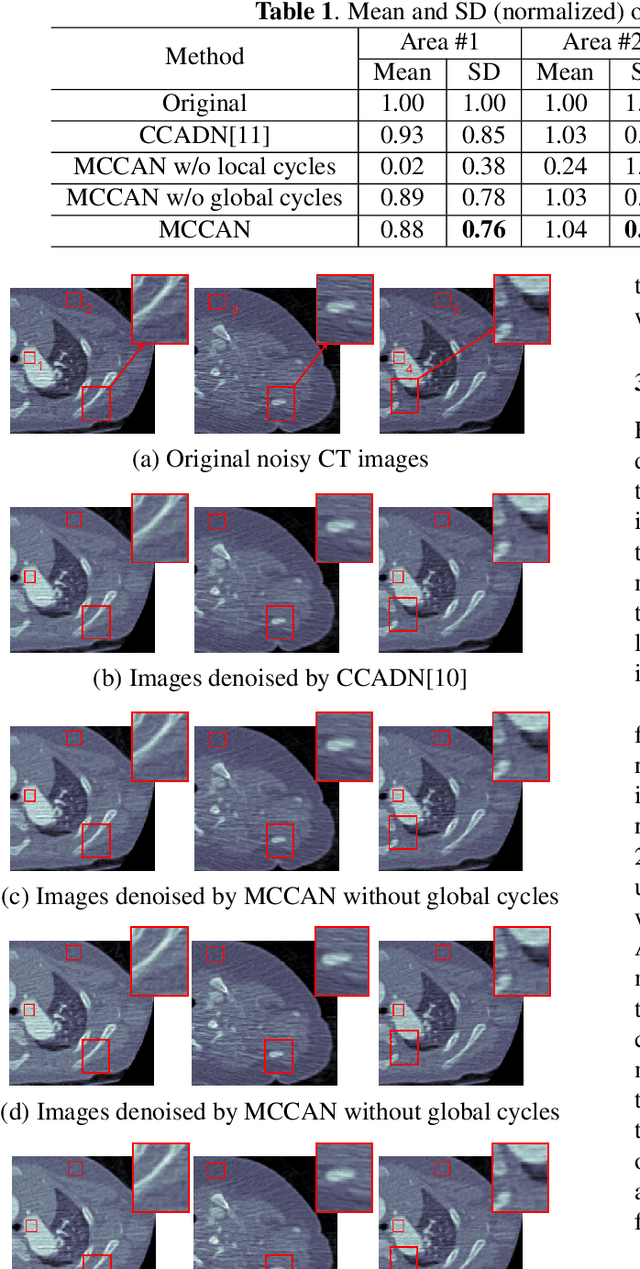
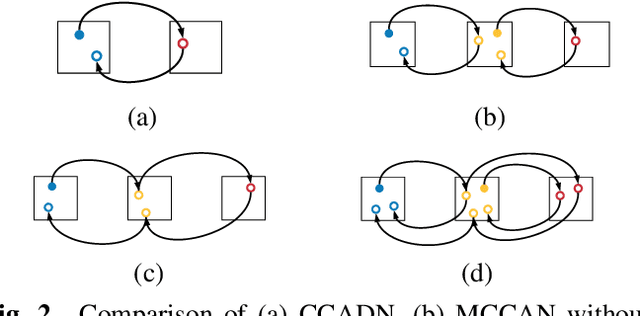
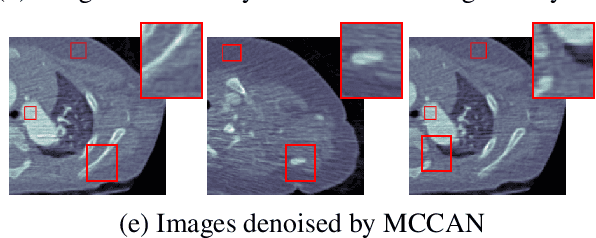
Abstract:CT image denoising can be treated as an image-to-image translation task where the goal is to learn the transform between a source domain $X$ (noisy images) and a target domain $Y$ (clean images). Recently, cycle-consistent adversarial denoising network (CCADN) has achieved state-of-the-art results by enforcing cycle-consistent loss without the need of paired training data. Our detailed analysis of CCADN raises a number of interesting questions. For example, if the noise is large leading to significant difference between domain $X$ and domain $Y$, can we bridge $X$ and $Y$ with an intermediate domain $Z$ such that both the denoising process between $X$ and $Z$ and that between $Z$ and $Y$ are easier to learn? As such intermediate domains lead to multiple cycles, how do we best enforce cycle-consistency? Driven by these questions, we propose a multi-cycle-consistent adversarial network (MCCAN) that builds intermediate domains and enforces both local and global cycle-consistency. The global cycle-consistency couples all generators together to model the whole denoising process, while the local cycle-consistency imposes effective supervision on the process between adjacent domains. Experiments show that both local and global cycle-consistency are important for the success of MCCAN, which outperforms the state-of-the-art.
MSU-Net: Multiscale Statistical U-Net for Real-time 3D Cardiac MRI Video Segmentation
Sep 15, 2019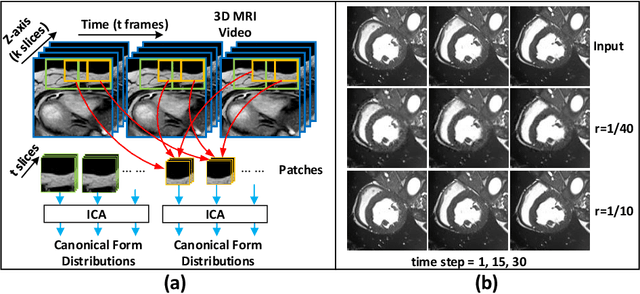
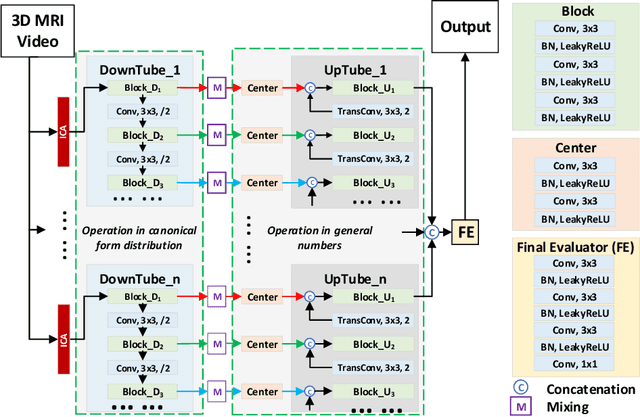
Abstract:Cardiac magnetic resonance imaging (MRI) is an essential tool for MRI-guided surgery and real-time intervention. The MRI videos are expected to be segmented on-the-fly in real practice. However, existing segmentation methods would suffer from drastic accuracy loss when modified for speedup. In this work, we propose Multiscale Statistical U-Net (MSU-Net) for real-time 3D MRI video segmentation in cardiac surgical guidance. Our idea is to model the input samples as multiscale canonical form distributions for speedup, while the spatio-temporal correlation is still fully utilized. A parallel statistical U-Net is then designed to efficiently process these distributions. The fast data sampling and efficient parallel structure of MSU-Net endorse the fast and accurate inference. Compared with vanilla U-Net and a modified state-of-the-art method GridNet, our method achieves up to 268% and 237% speedup with 1.6% and 3.6% increased Dice scores.
Accurate Congenital Heart Disease Model Generation for 3D Printing
Jul 12, 2019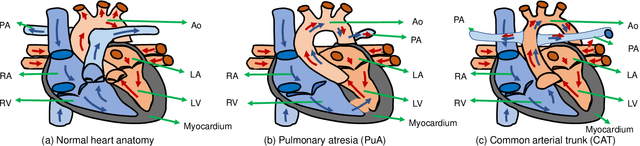
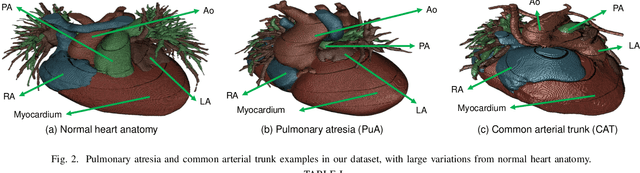
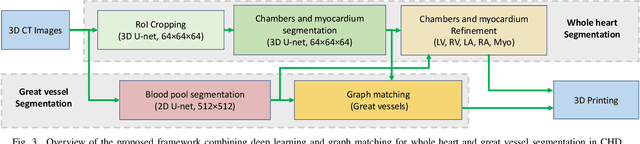
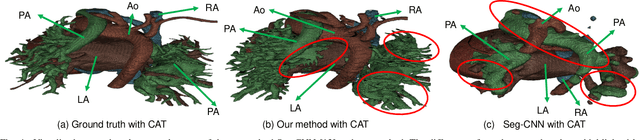
Abstract:3D printing has been widely adopted for clinical decision making and interventional planning of Congenital heart disease (CHD), while whole heart and great vessel segmentation is the most significant but time-consuming step in the model generation for 3D printing. While various automatic whole heart and great vessel segmentation frameworks have been developed in the literature, they are ineffective when applied to medical images in CHD, which have significant variations in heart structure and great vessel connections. To address the challenge, we leverage the power of deep learning in processing regular structures and that of graph algorithms in dealing with large variations and propose a framework that combines both for whole heart and great vessel segmentation in CHD. Particularly, we first use deep learning to segment the four chambers and myocardium followed by the blood pool, where variations are usually small. We then extract the connection information and apply graph matching to determine the categories of all the vessels. Experimental results using 683D CT images covering 14 types of CHD show that our method can increase Dice score by 11.9% on average compared with the state-of-the-art whole heart and great vessel segmentation method in normal anatomy. The segmentation results are also printed out using 3D printers for validation.
Machine Vision Guided 3D Medical Image Compression for Efficient Transmission and Accurate Segmentation in the Clouds
Apr 09, 2019

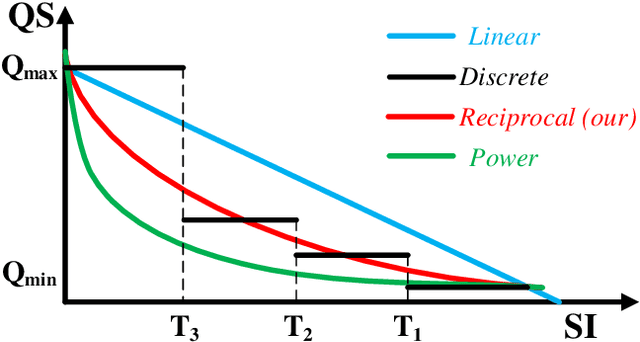
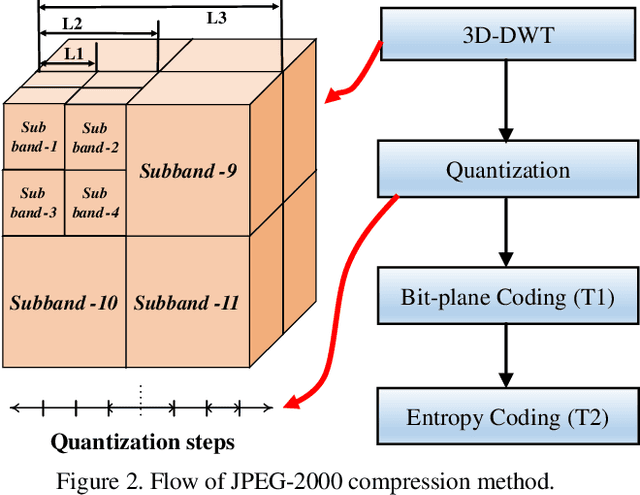
Abstract:Cloud based medical image analysis has become popular recently due to the high computation complexities of various deep neural network (DNN) based frameworks and the increasingly large volume of medical images that need to be processed. It has been demonstrated that for medical images the transmission from local to clouds is much more expensive than the computation in the clouds itself. Towards this, 3D image compression techniques have been widely applied to reduce the data traffic. However, most of the existing image compression techniques are developed around human vision, i.e., they are designed to minimize distortions that can be perceived by human eyes. In this paper we will use deep learning based medical image segmentation as a vehicle and demonstrate that interestingly, machine and human view the compression quality differently. Medical images compressed with good quality w.r.t. human vision may result in inferior segmentation accuracy. We then design a machine vision oriented 3D image compression framework tailored for segmentation using DNNs. Our method automatically extracts and retains image features that are most important to the segmentation. Comprehensive experiments on widely adopted segmentation frameworks with HVSMR 2016 challenge dataset show that our method can achieve significantly higher segmentation accuracy at the same compression rate, or much better compression rate under the same segmentation accuracy, when compared with the existing JPEG 2000 method. To the best of the authors' knowledge, this is the first machine vision guided medical image compression framework for segmentation in the clouds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge