Jaime S. Cardoso
Second Edition FRCSyn Challenge at CVPR 2024: Face Recognition Challenge in the Era of Synthetic Data
Apr 16, 2024



Abstract:Synthetic data is gaining increasing relevance for training machine learning models. This is mainly motivated due to several factors such as the lack of real data and intra-class variability, time and errors produced in manual labeling, and in some cases privacy concerns, among others. This paper presents an overview of the 2nd edition of the Face Recognition Challenge in the Era of Synthetic Data (FRCSyn) organized at CVPR 2024. FRCSyn aims to investigate the use of synthetic data in face recognition to address current technological limitations, including data privacy concerns, demographic biases, generalization to novel scenarios, and performance constraints in challenging situations such as aging, pose variations, and occlusions. Unlike the 1st edition, in which synthetic data from DCFace and GANDiffFace methods was only allowed to train face recognition systems, in this 2nd edition we propose new sub-tasks that allow participants to explore novel face generative methods. The outcomes of the 2nd FRCSyn Challenge, along with the proposed experimental protocol and benchmarking contribute significantly to the application of synthetic data to face recognition.
* arXiv admin note: text overlap with arXiv:2311.10476
Anonymizing medical case-based explanations through disentanglement
Nov 08, 2023



Abstract:Case-based explanations are an intuitive method to gain insight into the decision-making process of deep learning models in clinical contexts. However, medical images cannot be shared as explanations due to privacy concerns. To address this problem, we propose a novel method for disentangling identity and medical characteristics of images and apply it to anonymize medical images. The disentanglement mechanism replaces some feature vectors in an image while ensuring that the remaining features are preserved, obtaining independent feature vectors that encode the images' identity and medical characteristics. We also propose a model to manufacture synthetic privacy-preserving identities to replace the original image's identity and achieve anonymization. The models are applied to medical and biometric datasets, demonstrating their capacity to generate realistic-looking anonymized images that preserve their original medical content. Additionally, the experiments show the network's inherent capacity to generate counterfactual images through the replacement of medical features.
Compressed Models Decompress Race Biases: What Quantized Models Forget for Fair Face Recognition
Aug 23, 2023


Abstract:With the ever-growing complexity of deep learning models for face recognition, it becomes hard to deploy these systems in real life. Researchers have two options: 1) use smaller models; 2) compress their current models. Since the usage of smaller models might lead to concerning biases, compression gains relevance. However, compressing might be also responsible for an increase in the bias of the final model. We investigate the overall performance, the performance on each ethnicity subgroup and the racial bias of a State-of-the-Art quantization approach when used with synthetic and real data. This analysis provides a few more details on potential benefits of performing quantization with synthetic data, for instance, the reduction of biases on the majority of test scenarios. We tested five distinct architectures and three different training datasets. The models were evaluated on a fourth dataset which was collected to infer and compare the performance of face recognition models on different ethnicity.
Unveiling the Two-Faced Truth: Disentangling Morphed Identities for Face Morphing Detection
Jun 05, 2023

Abstract:Morphing attacks keep threatening biometric systems, especially face recognition systems. Over time they have become simpler to perform and more realistic, as such, the usage of deep learning systems to detect these attacks has grown. At the same time, there is a constant concern regarding the lack of interpretability of deep learning models. Balancing performance and interpretability has been a difficult task for scientists. However, by leveraging domain information and proving some constraints, we have been able to develop IDistill, an interpretable method with state-of-the-art performance that provides information on both the identity separation on morph samples and their contribution to the final prediction. The domain information is learnt by an autoencoder and distilled to a classifier system in order to teach it to separate identity information. When compared to other methods in the literature it outperforms them in three out of five databases and is competitive in the remaining.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Unimodal Distributions for Ordinal Regression
Mar 08, 2023Abstract:In many real-world prediction tasks, class labels contain information about the relative order between labels that are not captured by commonly used loss functions such as multicategory cross-entropy. Recently, the preference for unimodal distributions in the output space has been incorporated into models and loss functions to account for such ordering information. However, current approaches rely on heuristics that lack a theoretical foundation. Here, we propose two new approaches to incorporate the preference for unimodal distributions into the predictive model. We analyse the set of unimodal distributions in the probability simplex and establish fundamental properties. We then propose a new architecture that imposes unimodal distributions and a new loss term that relies on the notion of projection in a set to promote unimodality. Experiments show the new architecture achieves top-2 performance, while the proposed new loss term is very competitive while maintaining high unimodality.
A CAD System for Colorectal Cancer from WSI: A Clinically Validated Interpretable ML-based Prototype
Jan 06, 2023Abstract:The integration of Artificial Intelligence (AI) and Digital Pathology has been increasing over the past years. Nowadays, applications of deep learning (DL) methods to diagnose cancer from whole-slide images (WSI) are, more than ever, a reality within different research groups. Nonetheless, the development of these systems was limited by a myriad of constraints regarding the lack of training samples, the scaling difficulties, the opaqueness of DL methods, and, more importantly, the lack of clinical validation. As such, we propose a system designed specifically for the diagnosis of colorectal samples. The construction of such a system consisted of four stages: (1) a careful data collection and annotation process, which resulted in one of the largest WSI colorectal samples datasets; (2) the design of an interpretable mixed-supervision scheme to leverage the domain knowledge introduced by pathologists through spatial annotations; (3) the development of an effective sampling approach based on the expected severeness of each tile, which decreased the computation cost by a factor of almost 6x; (4) the creation of a prototype that integrates the full set of features of the model to be evaluated in clinical practice. During these stages, the proposed method was evaluated in four separate test sets, two of them are external and completely independent. On the largest of those sets, the proposed approach achieved an accuracy of 93.44%. DL for colorectal samples is a few steps closer to stop being research exclusive and to become fully integrated in clinical practice.
PIC-Score: Probabilistic Interpretable Comparison Score for Optimal Matching Confidence in Single- and Multi-Biometric (Face) Recognition
Nov 22, 2022



Abstract:In the context of biometrics, matching confidence refers to the confidence that a given matching decision is correct. Since many biometric systems operate in critical decision-making processes, such as in forensics investigations, accurately and reliably stating the matching confidence becomes of high importance. Previous works on biometric confidence estimation can well differentiate between high and low confidence, but lack interpretability. Therefore, they do not provide accurate probabilistic estimates of the correctness of a decision. In this work, we propose a probabilistic interpretable comparison (PIC) score that accurately reflects the probability that the score originates from samples of the same identity. We prove that the proposed approach provides optimal matching confidence. Contrary to other approaches, it can also optimally combine multiple samples in a joint PIC score which further increases the recognition and confidence estimation performance. In the experiments, the proposed PIC approach is compared against all biometric confidence estimation methods available on four publicly available databases and five state-of-the-art face recognition systems. The results demonstrate that PIC has a significantly more accurate probabilistic interpretation than similar approaches and is highly effective for multi-biometric recognition. The code is publicly-available.
OrthoMAD: Morphing Attack Detection Through Orthogonal Identity Disentanglement
Aug 23, 2022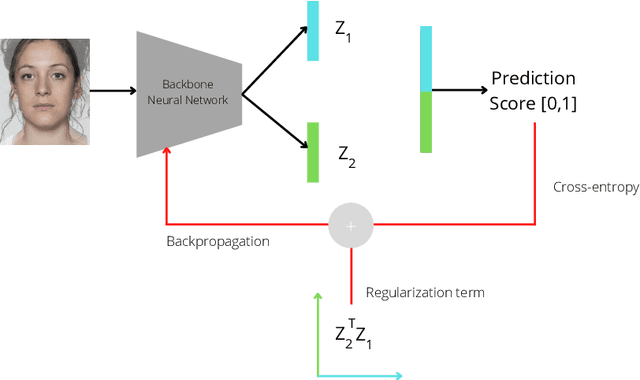
Abstract:Morphing attacks are one of the many threats that are constantly affecting deep face recognition systems. It consists of selecting two faces from different individuals and fusing them into a final image that contains the identity information of both. In this work, we propose a novel regularisation term that takes into account the existent identity information in both and promotes the creation of two orthogonal latent vectors. We evaluate our proposed method (OrthoMAD) in five different types of morphing in the FRLL dataset and evaluate the performance of our model when trained on five distinct datasets. With a small ResNet-18 as the backbone, we achieve state-of-the-art results in the majority of the experiments, and competitive results in the others. The code of this paper will be publicly available.
Explainable Biometrics in the Age of Deep Learning
Aug 19, 2022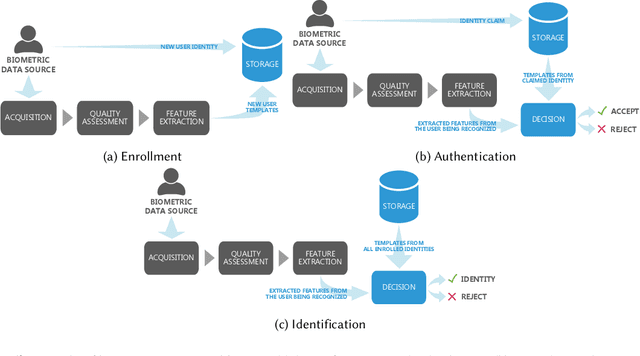
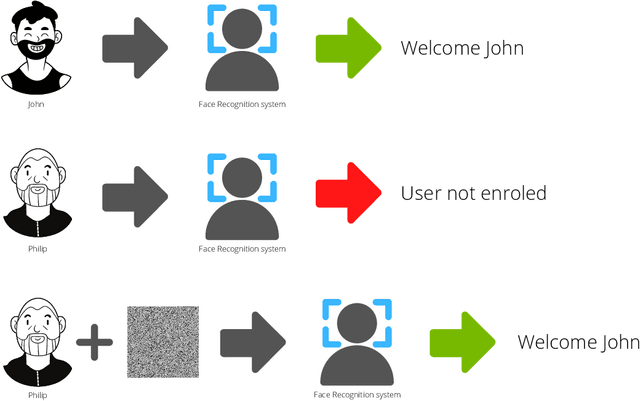
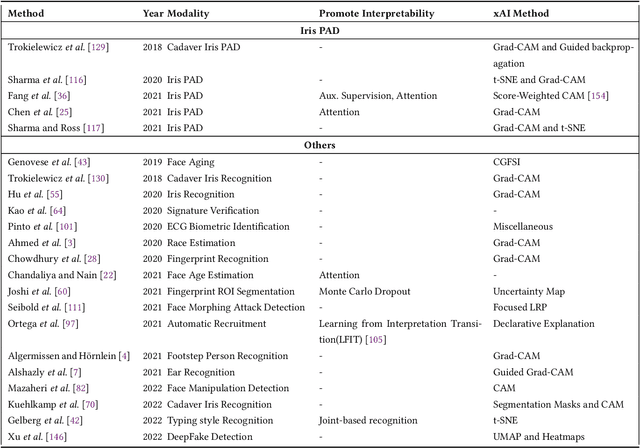
Abstract:Systems capable of analyzing and quantifying human physical or behavioral traits, known as biometrics systems, are growing in use and application variability. Since its evolution from handcrafted features and traditional machine learning to deep learning and automatic feature extraction, the performance of biometric systems increased to outstanding values. Nonetheless, the cost of this fast progression is still not understood. Due to its opacity, deep neural networks are difficult to understand and analyze, hence, hidden capacities or decisions motivated by the wrong motives are a potential risk. Researchers have started to pivot their focus towards the understanding of deep neural networks and the explanation of their predictions. In this paper, we provide a review of the current state of explainable biometrics based on the study of 47 papers and discuss comprehensively the direction in which this field should be developed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge