Huimin Cheng
DCMM-Transformer: Degree-Corrected Mixed-Membership Attention for Medical Imaging
Nov 15, 2025Abstract:Medical images exhibit latent anatomical groupings, such as organs, tissues, and pathological regions, that standard Vision Transformers (ViTs) fail to exploit. While recent work like SBM-Transformer attempts to incorporate such structures through stochastic binary masking, they suffer from non-differentiability, training instability, and the inability to model complex community structure. We present DCMM-Transformer, a novel ViT architecture for medical image analysis that incorporates a Degree-Corrected Mixed-Membership (DCMM) model as an additive bias in self-attention. Unlike prior approaches that rely on multiplicative masking and binary sampling, our method introduces community structure and degree heterogeneity in a fully differentiable and interpretable manner. Comprehensive experiments across diverse medical imaging datasets, including brain, chest, breast, and ocular modalities, demonstrate the superior performance and generalizability of the proposed approach. Furthermore, the learned group structure and structured attention modulation substantially enhance interpretability by yielding attention maps that are anatomically meaningful and semantically coherent.
Class-balanced Open-set Semi-supervised Object Detection for Medical Images
Aug 22, 2024



Abstract:Medical image datasets in the real world are often unlabeled and imbalanced, and Semi-Supervised Object Detection (SSOD) can utilize unlabeled data to improve an object detector. However, existing approaches predominantly assumed that the unlabeled data and test data do not contain out-of-distribution (OOD) classes. The few open-set semi-supervised object detection methods have two weaknesses: first, the class imbalance is not considered; second, the OOD instances are distinguished and simply discarded during pseudo-labeling. In this paper, we consider the open-set semi-supervised object detection problem which leverages unlabeled data that contain OOD classes to improve object detection for medical images. Our study incorporates two key innovations: Category Control Embed (CCE) and out-of-distribution Detection Fusion Classifier (OODFC). CCE is designed to tackle dataset imbalance by constructing a Foreground information Library, while OODFC tackles open-set challenges by integrating the ``unknown'' information into basic pseudo-labels. Our method outperforms the state-of-the-art SSOD performance, achieving a 4.25 mAP improvement on the public Parasite dataset.
Ensemble machine learning approach for screening of coronary heart disease based on echocardiography and risk factors
May 20, 2021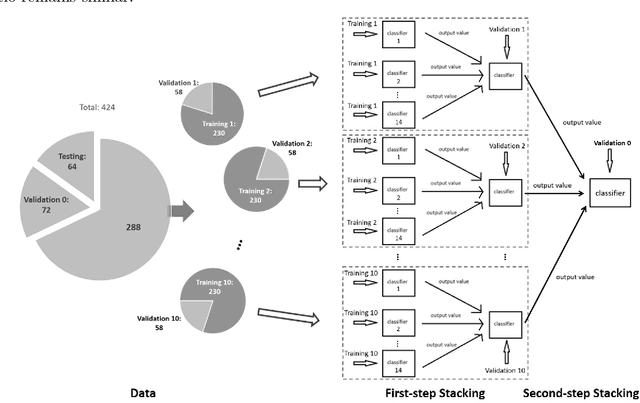
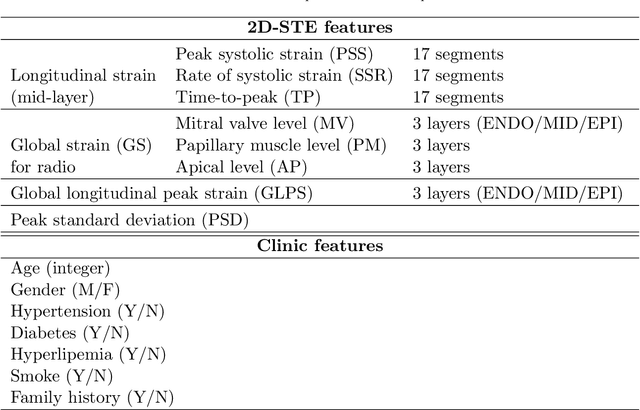
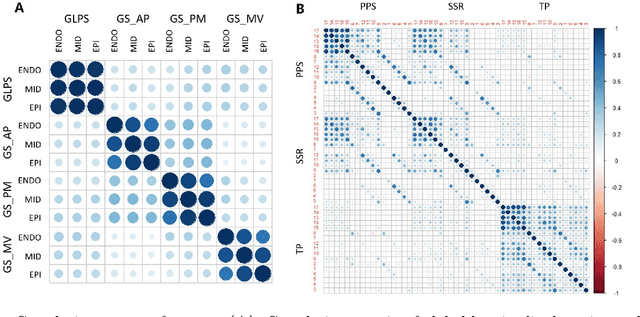
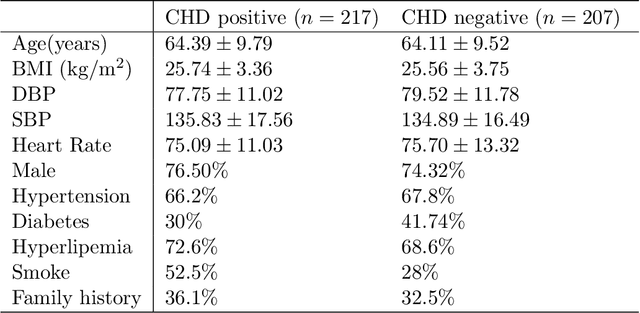
Abstract:Background: Extensive clinical evidence suggests that a preventive screening of coronary heart disease (CHD) at an earlier stage can greatly reduce the mortality rate. We use 64 two-dimensional speckle tracking echocardiography (2D-STE) features and seven clinical features to predict whether one has CHD. Methods: We develop a machine learning approach that integrates a number of popular classification methods together by model stacking, and generalize the traditional stacking method to a two-step stacking method to improve the diagnostic performance. Results: By borrowing strengths from multiple classification models through the proposed method, we improve the CHD classification accuracy from around 70% to 87.7% on the testing set. The sensitivity of the proposed method is 0.903 and the specificity is 0.843, with an AUC of 0.904, which is significantly higher than those of the individual classification models. Conclusions: Our work lays a foundation for the deployment of speckle tracking echocardiography-based screening tools for coronary heart disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge