Yongkai Chen
Classifying Metamorphic versus Single-Fold Proteins with Statistical Learning and AlphaFold2
Dec 10, 2025Abstract:The remarkable success of AlphaFold2 in providing accurate atomic-level prediction of protein structures from their amino acid sequence has transformed approaches to the protein folding problem. However, its core paradigm of mapping one sequence to one structure may only be appropriate for single-fold proteins with one stable conformation. Metamorphic proteins, which can adopt multiple distinct conformations, have conformational diversity that cannot be adequately modeled by AlphaFold2. Hence, classifying whether a given protein is metamorphic or single-fold remains a critical challenge for both laboratory experiments and computational methods. To address this challenge, we developed a novel classification framework by re-purposing AlphaFold2 to generate conformational ensembles via a multiple sequence alignment sampling method. From these ensembles, we extract a comprehensive set of features characterizing the conformational ensemble's modality and structural dispersion. A random forest classifier trained on a carefully curated benchmark dataset of known metamorphic and single-fold proteins achieves a mean AUC of 0.869 with cross-validation, demonstrating the effectiveness of our integrated approach. Furthermore, by applying our classifier to 600 randomly sampled proteins from the Protein Data Bank, we identified several potential metamorphic protein candidates -- including the 40S ribosomal protein S30, whose conformational change is crucial for its secondary function in antimicrobial defense. By combining AI-driven protein structure prediction with statistical learning, our work provides a powerful new approach for discovering metamorphic proteins and deepens our understanding of their role in their molecular function.
S2MNet: Speckle-To-Mesh Net for Three-Dimensional Cardiac Morphology Reconstruction via Echocardiogram
May 09, 2025Abstract:Echocardiogram is the most commonly used imaging modality in cardiac assessment duo to its non-invasive nature, real-time capability, and cost-effectiveness. Despite its advantages, most clinical echocardiograms provide only two-dimensional views, limiting the ability to fully assess cardiac anatomy and function in three dimensions. While three-dimensional echocardiography exists, it often suffers from reduced resolution, limited availability, and higher acquisition costs. To overcome these challenges, we propose a deep learning framework S2MNet that reconstructs continuous and high-fidelity 3D heart models by integrating six slices of routinely acquired 2D echocardiogram views. Our method has three advantages. First, our method avoid the difficulties on training data acquasition by simulate six of 2D echocardiogram images from corresponding slices of a given 3D heart mesh. Second, we introduce a deformation field-based method, which avoid spatial discontinuities or structural artifacts in 3D echocardiogram reconstructions. We validate our method using clinically collected echocardiogram and demonstrate that our estimated left ventricular volume, a key clinical indicator of cardiac function, is strongly correlated with the doctor measured GLPS, a clinical measurement that should demonstrate a negative correlation with LVE in medical theory. This association confirms the reliability of our proposed 3D construction method.
Knowledge Distillation and Dataset Distillation of Large Language Models: Emerging Trends, Challenges, and Future Directions
Apr 20, 2025



Abstract:The exponential growth of Large Language Models (LLMs) continues to highlight the need for efficient strategies to meet ever-expanding computational and data demands. This survey provides a comprehensive analysis of two complementary paradigms: Knowledge Distillation (KD) and Dataset Distillation (DD), both aimed at compressing LLMs while preserving their advanced reasoning capabilities and linguistic diversity. We first examine key methodologies in KD, such as task-specific alignment, rationale-based training, and multi-teacher frameworks, alongside DD techniques that synthesize compact, high-impact datasets through optimization-based gradient matching, latent space regularization, and generative synthesis. Building on these foundations, we explore how integrating KD and DD can produce more effective and scalable compression strategies. Together, these approaches address persistent challenges in model scalability, architectural heterogeneity, and the preservation of emergent LLM abilities. We further highlight applications across domains such as healthcare and education, where distillation enables efficient deployment without sacrificing performance. Despite substantial progress, open challenges remain in preserving emergent reasoning and linguistic diversity, enabling efficient adaptation to continually evolving teacher models and datasets, and establishing comprehensive evaluation protocols. By synthesizing methodological innovations, theoretical foundations, and practical insights, our survey charts a path toward sustainable, resource-efficient LLMs through the tighter integration of KD and DD principles.
Ensemble machine learning approach for screening of coronary heart disease based on echocardiography and risk factors
May 20, 2021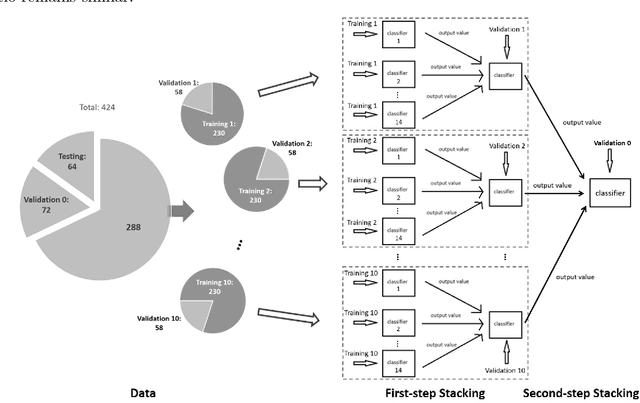
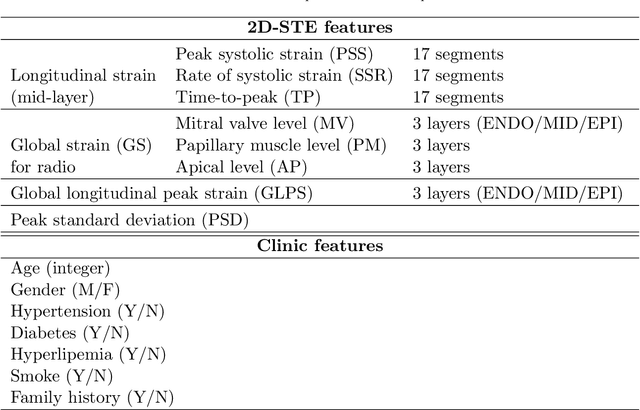
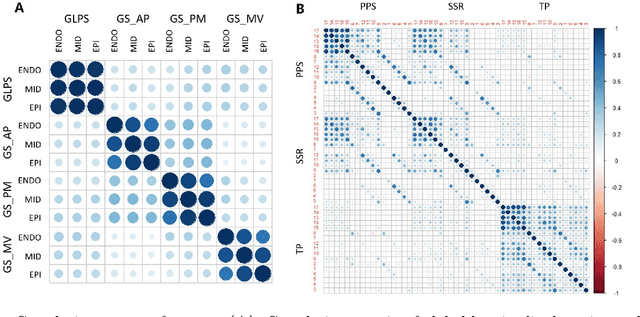
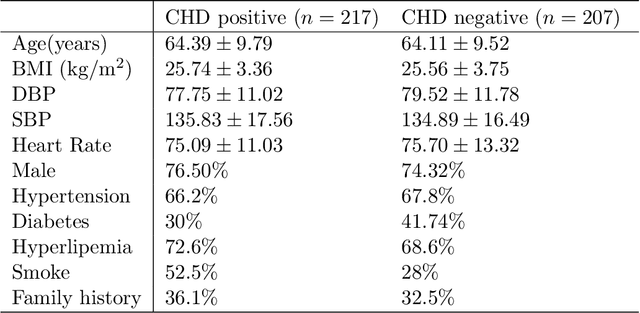
Abstract:Background: Extensive clinical evidence suggests that a preventive screening of coronary heart disease (CHD) at an earlier stage can greatly reduce the mortality rate. We use 64 two-dimensional speckle tracking echocardiography (2D-STE) features and seven clinical features to predict whether one has CHD. Methods: We develop a machine learning approach that integrates a number of popular classification methods together by model stacking, and generalize the traditional stacking method to a two-step stacking method to improve the diagnostic performance. Results: By borrowing strengths from multiple classification models through the proposed method, we improve the CHD classification accuracy from around 70% to 87.7% on the testing set. The sensitivity of the proposed method is 0.903 and the specificity is 0.843, with an AUC of 0.904, which is significantly higher than those of the individual classification models. Conclusions: Our work lays a foundation for the deployment of speckle tracking echocardiography-based screening tools for coronary heart disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge