Hoo-Chang Shin
HelpSteer3-Preference: Open Human-Annotated Preference Data across Diverse Tasks and Languages
May 16, 2025Abstract:Preference datasets are essential for training general-domain, instruction-following language models with Reinforcement Learning from Human Feedback (RLHF). Each subsequent data release raises expectations for future data collection, meaning there is a constant need to advance the quality and diversity of openly available preference data. To address this need, we introduce HelpSteer3-Preference, a permissively licensed (CC-BY-4.0), high-quality, human-annotated preference dataset comprising of over 40,000 samples. These samples span diverse real-world applications of large language models (LLMs), including tasks relating to STEM, coding and multilingual scenarios. Using HelpSteer3-Preference, we train Reward Models (RMs) that achieve top performance on RM-Bench (82.4%) and JudgeBench (73.7%). This represents a substantial improvement (~10% absolute) over the previously best-reported results from existing RMs. We demonstrate HelpSteer3-Preference can also be applied to train Generative RMs and how policy models can be aligned with RLHF using our RMs. Dataset (CC-BY-4.0): https://huggingface.co/datasets/nvidia/HelpSteer3#preference
Dedicated Feedback and Edit Models Empower Inference-Time Scaling for Open-Ended General-Domain Tasks
Mar 06, 2025



Abstract:Inference-Time Scaling has been critical to the success of recent models such as OpenAI o1 and DeepSeek R1. However, many techniques used to train models for inference-time scaling require tasks to have answers that can be verified, limiting their application to domains such as math, coding and logical reasoning. We take inspiration from how humans make first attempts, ask for detailed feedback from others and make improvements based on such feedback across a wide spectrum of open-ended endeavors. To this end, we collect data for and train dedicated Feedback and Edit Models that are capable of performing inference-time scaling for open-ended general-domain tasks. In our setup, one model generates an initial response, which are given feedback by a second model, that are then used by a third model to edit the response. We show that performance on Arena Hard, a benchmark strongly predictive of Chatbot Arena Elo can be boosted by scaling the number of initial response drafts, effective feedback and edited responses. When scaled optimally, our setup based on 70B models from the Llama 3 family can reach SoTA performance on Arena Hard at 92.7 as of 5 Mar 2025, surpassing OpenAI o1-preview-2024-09-12 with 90.4 and DeepSeek R1 with 92.3.
Automatic Extraction of Medication Names in Tweets as Named Entity Recognition
Nov 30, 2021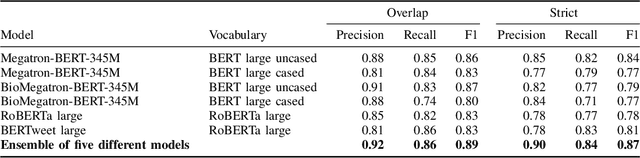
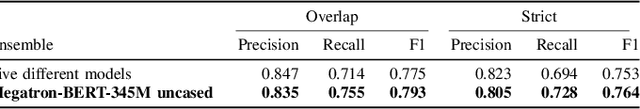
Abstract:Social media posts contain potentially valuable information about medical conditions and health-related behavior. Biocreative VII Task 3 focuses on mining this information by recognizing mentions of medications and dietary supplements in tweets. We approach this task by fine tuning multiple BERT-style language models to perform token-level classification, and combining them into ensembles to generate final predictions. Our best system consists of five Megatron-BERT-345M models and achieves a strict F1 score of 0.764 on unseen test data.
Chemical Identification and Indexing in PubMed Articles via BERT and Text-to-Text Approaches
Nov 30, 2021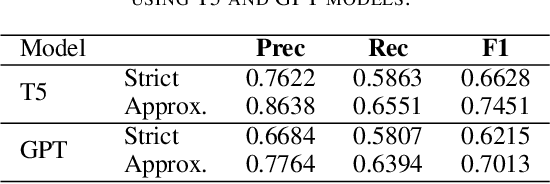

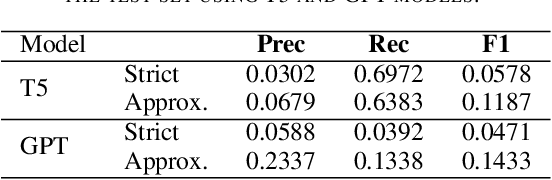
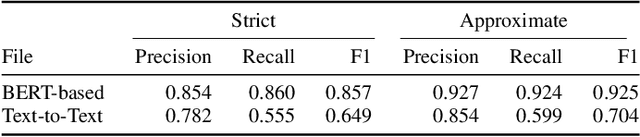
Abstract:The Biocreative VII Track-2 challenge consists of named entity recognition, entity-linking (or entity-normalization), and topic indexing tasks -- with entities and topics limited to chemicals for this challenge. Named entity recognition is a well-established problem and we achieve our best performance with BERT-based BioMegatron models. We extend our BERT-based approach to the entity linking task. After the second stage of pretraining BioBERT with a metric-learning loss strategy called self-alignment pretraining (SAP), we link entities based on the cosine similarity between their SAP-BioBERT word embeddings. Despite the success of our named entity recognition experiments, we find the chemical indexing task generally more challenging. In addition to conventional NER methods, we attempt both named entity recognition and entity linking with a novel text-to-text or "prompt" based method that uses generative language models such as T5 and GPT. We achieve encouraging results with this new approach.
Text Mining Drug/Chemical-Protein Interactions using an Ensemble of BERT and T5 Based Models
Nov 30, 2021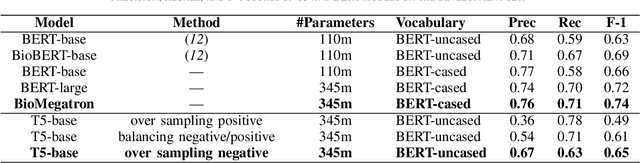

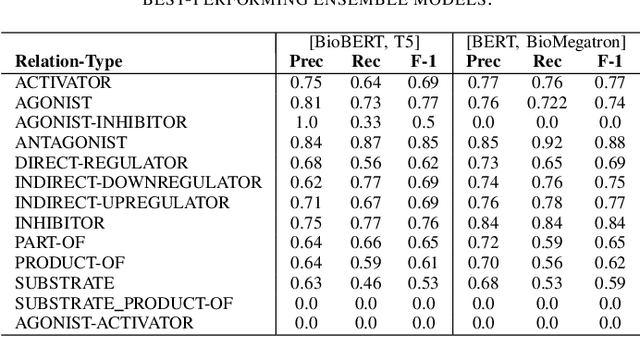
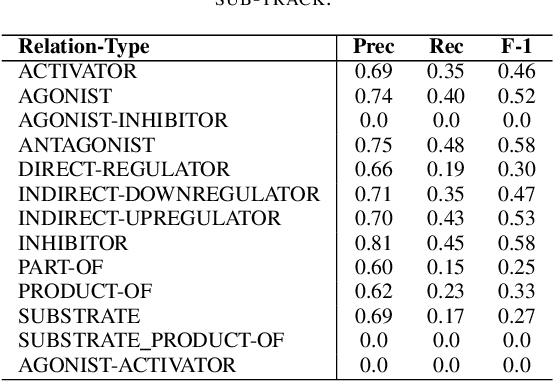
Abstract:In Track-1 of the BioCreative VII Challenge participants are asked to identify interactions between drugs/chemicals and proteins. In-context named entity annotations for each drug/chemical and protein are provided and one of fourteen different interactions must be automatically predicted. For this relation extraction task, we attempt both a BERT-based sentence classification approach, and a more novel text-to-text approach using a T5 model. We find that larger BERT-based models perform better in general, with our BioMegatron-based model achieving the highest scores across all metrics, achieving 0.74 F1 score. Though our novel T5 text-to-text method did not perform as well as most of our BERT-based models, it outperformed those trained on similar data, showing promising results, achieving 0.65 F1 score. We believe a text-to-text approach to relation extraction has some competitive advantages and there is a lot of room for research advancement.
BioMegatron: Larger Biomedical Domain Language Model
Oct 14, 2020
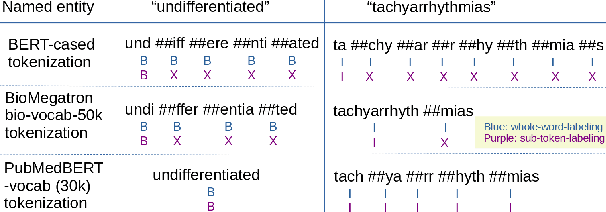
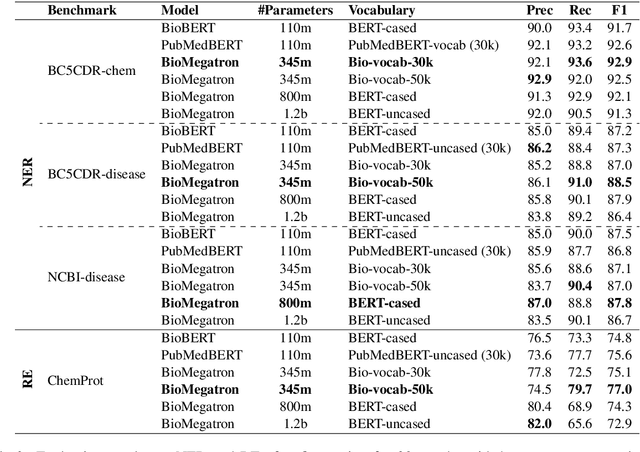
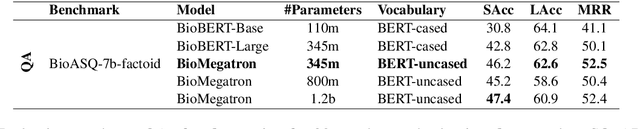
Abstract:There has been an influx of biomedical domain-specific language models, showing language models pre-trained on biomedical text perform better on biomedical domain benchmarks than those trained on general domain text corpora such as Wikipedia and Books. Yet, most works do not study the factors affecting each domain language application deeply. Additionally, the study of model size on domain-specific models has been mostly missing. We empirically study and evaluate several factors that can affect performance on domain language applications, such as the sub-word vocabulary set, model size, pre-training corpus, and domain transfer. We show consistent improvements on benchmarks with our larger BioMegatron model trained on a larger domain corpus, contributing to our understanding of domain language model applications. We demonstrate noticeable improvements over the previous state-of-the-art (SOTA) on standard biomedical NLP benchmarks of named entity recognition, relation extraction, and question answering. Model checkpoints and code are available at [https://ngc.nvidia.com] and [https://github.com/NVIDIA/NeMo].
GANDALF: Generative Adversarial Networks with Discriminator-Adaptive Loss Fine-tuning for Alzheimer's Disease Diagnosis from MRI
Aug 10, 2020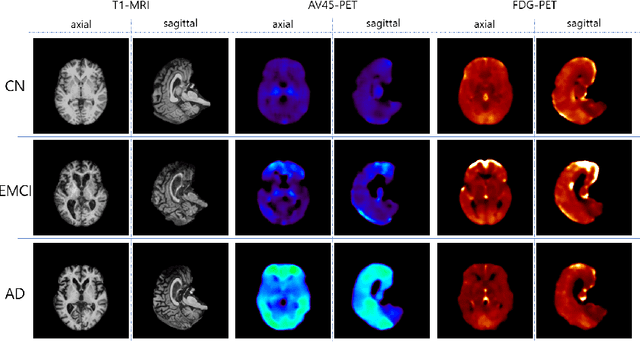
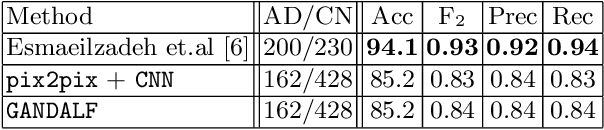
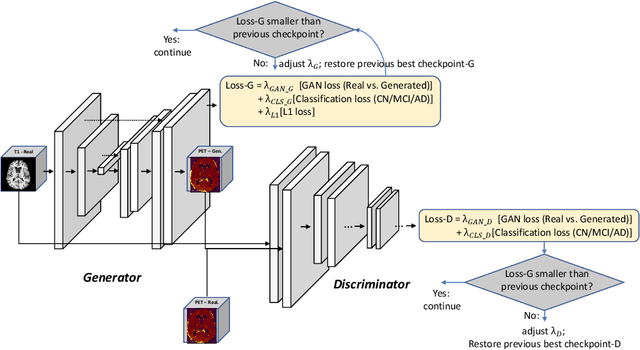
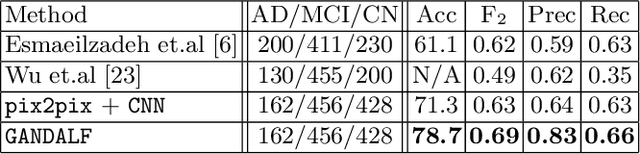
Abstract:Positron Emission Tomography (PET) is now regarded as the gold standard for the diagnosis of Alzheimer's Disease (AD). However, PET imaging can be prohibitive in terms of cost and planning, and is also among the imaging techniques with the highest dosage of radiation. Magnetic Resonance Imaging (MRI), in contrast, is more widely available and provides more flexibility when setting the desired image resolution. Unfortunately, the diagnosis of AD using MRI is difficult due to the very subtle physiological differences between healthy and AD subjects visible on MRI. As a result, many attempts have been made to synthesize PET images from MR images using generative adversarial networks (GANs) in the interest of enabling the diagnosis of AD from MR. Existing work on PET synthesis from MRI has largely focused on Conditional GANs, where MR images are used to generate PET images and subsequently used for AD diagnosis. There is no end-to-end training goal. This paper proposes an alternative approach to the aforementioned, where AD diagnosis is incorporated in the GAN training objective to achieve the best AD classification performance. Different GAN lossesare fine-tuned based on the discriminator performance, and the overall training is stabilized. The proposed network architecture and training regime show state-of-the-art performance for three- and four- class AD classification tasks.
GANBERT: Generative Adversarial Networks with Bidirectional Encoder Representations from Transformers for MRI to PET synthesis
Aug 10, 2020
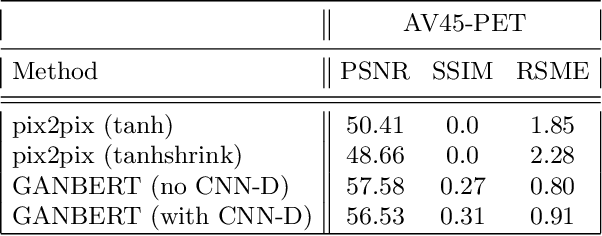
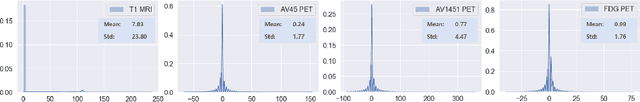
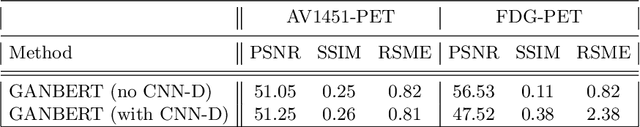
Abstract:Synthesizing medical images, such as PET, is a challenging task due to the fact that the intensity range is much wider and denser than those in photographs and digital renderings and are often heavily biased toward zero. Above all, intensity values in PET have absolute significance, and are used to compute parameters that are reproducible across the population. Yet, usually much manual adjustment has to be made in pre-/post- processing when synthesizing PET images, because its intensity ranges can vary a lot, e.g., between -100 to 1000 in floating point values. To overcome these challenges, we adopt the Bidirectional Encoder Representations from Transformers (BERT) algorithm that has had great success in natural language processing (NLP), where wide-range floating point intensity values are represented as integers ranging between 0 to 10000 that resemble a dictionary of natural language vocabularies. BERT is then trained to predict a proportion of masked values images, where its "next sentence prediction (NSP)" acts as GAN discriminator. Our proposed approach, is able to generate PET images from MRI images in wide intensity range, with no manual adjustments in pre-/post- processing. It is a method that can scale and ready to deploy.
Correlation via synthesis: end-to-end nodule image generation and radiogenomic map learning based on generative adversarial network
Jul 08, 2019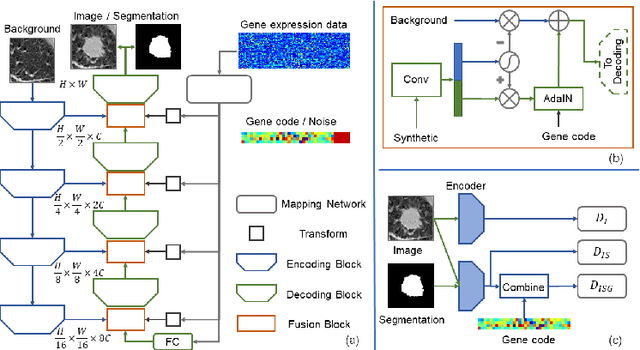

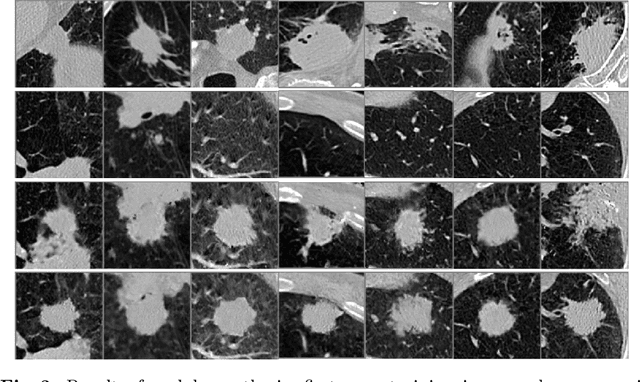
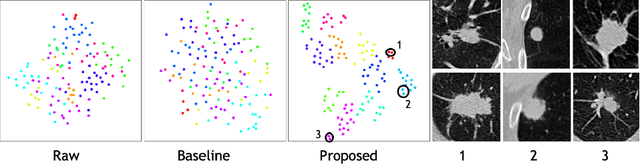
Abstract:Radiogenomic map linking image features and gene expression profiles is useful for noninvasively identifying molecular properties of a particular type of disease. Conventionally, such map is produced in three separate steps: 1) gene-clustering to "metagenes", 2) image feature extraction, and 3) statistical correlation between metagenes and image features. Each step is independently performed and relies on arbitrary measurements. In this work, we investigate the potential of an end-to-end method fusing gene data with image features to generate synthetic image and learn radiogenomic map simultaneously. To achieve this goal, we develop a generative adversarial network (GAN) conditioned on both background images and gene expression profiles, synthesizing the corresponding image. Image and gene features are fused at different scales to ensure the realism and quality of the synthesized image. We tested our method on non-small cell lung cancer (NSCLC) dataset. Results demonstrate that the proposed method produces realistic synthetic images, and provides a promising way to find gene-image relationship in a holistic end-to-end manner.
Medical Image Synthesis for Data Augmentation and Anonymization using Generative Adversarial Networks
Sep 13, 2018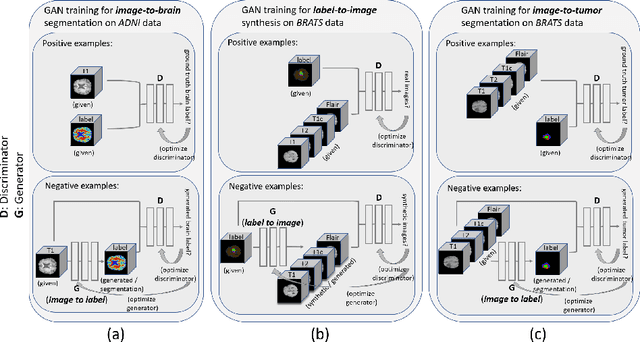
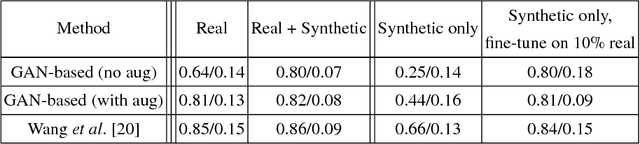
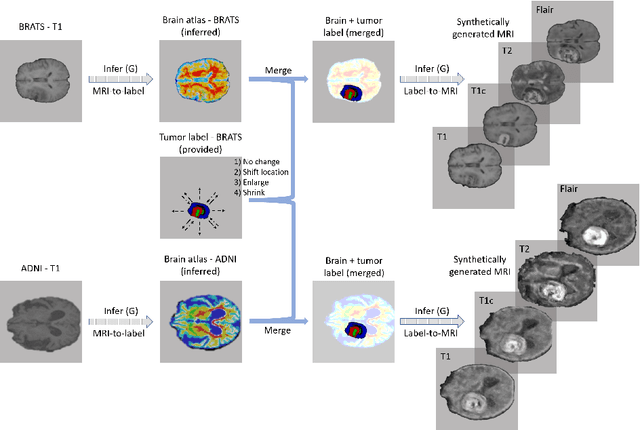
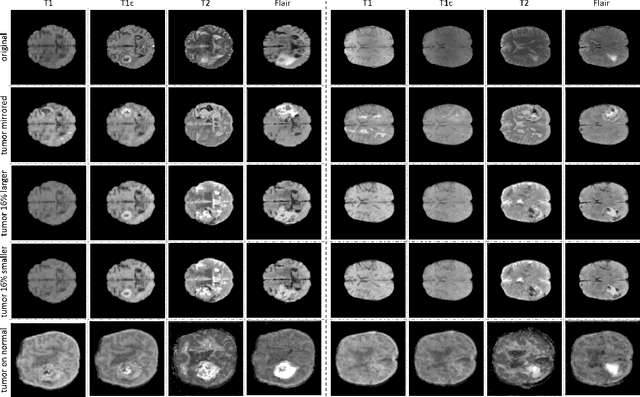
Abstract:Data diversity is critical to success when training deep learning models. Medical imaging data sets are often imbalanced as pathologic findings are generally rare, which introduces significant challenges when training deep learning models. In this work, we propose a method to generate synthetic abnormal MRI images with brain tumors by training a generative adversarial network using two publicly available data sets of brain MRI. We demonstrate two unique benefits that the synthetic images provide. First, we illustrate improved performance on tumor segmentation by leveraging the synthetic images as a form of data augmentation. Second, we demonstrate the value of generative models as an anonymization tool, achieving comparable tumor segmentation results when trained on the synthetic data versus when trained on real subject data. Together, these results offer a potential solution to two of the largest challenges facing machine learning in medical imaging, namely the small incidence of pathological findings, and the restrictions around sharing of patient data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge