Elodie Puybareau
EPITA Research Lab
Linear Object Detection in Document Images using Multiple Object Tracking
May 26, 2023



Abstract:Linear objects convey substantial information about document structure, but are challenging to detect accurately because of degradation (curved, erased) or decoration (doubled, dashed). Many approaches can recover some vector representation, but only one closed-source technique introduced in 1994, based on Kalman filters (a particular case of Multiple Object Tracking algorithm), can perform a pixel-accurate instance segmentation of linear objects and enable to selectively remove them from the original image. We aim at re-popularizing this approach and propose: 1. a framework for accurate instance segmentation of linear objects in document images using Multiple Object Tracking (MOT); 2. document image datasets and metrics which enable both vector- and pixel-based evaluation of linear object detection; 3. performance measures of MOT approaches against modern segment detectors; 4. performance measures of various tracking strategies, exhibiting alternatives to the original Kalman filters approach; and 5. an open-source implementation of a detector which can discriminate instances of curved, erased, dashed, intersecting and/or overlapping linear objects.
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022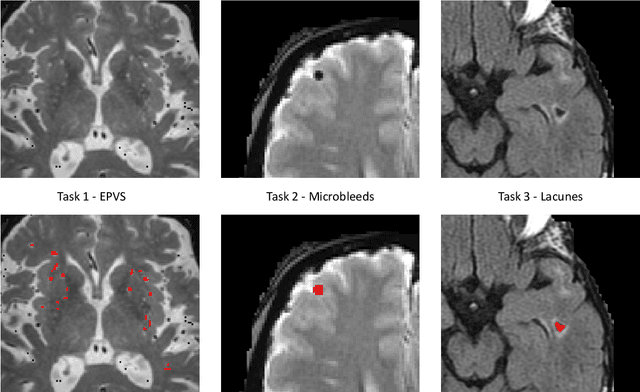
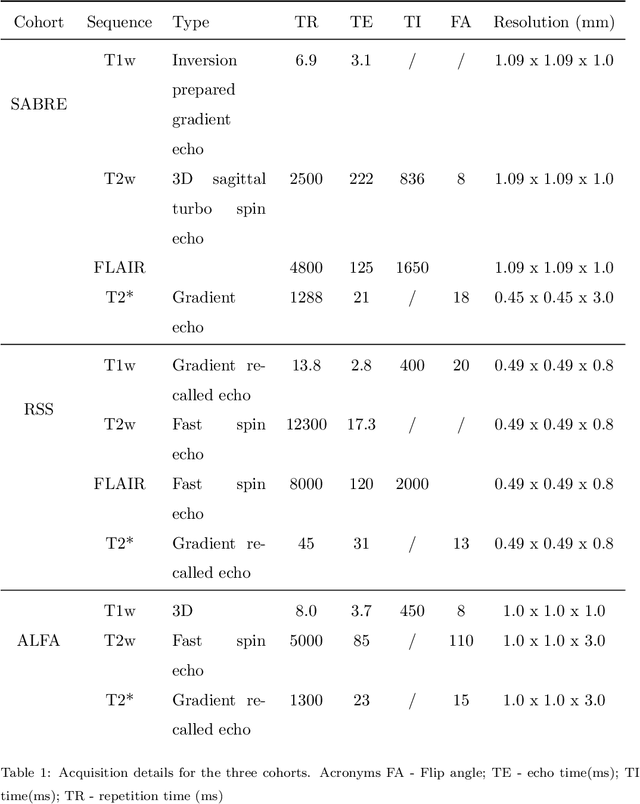
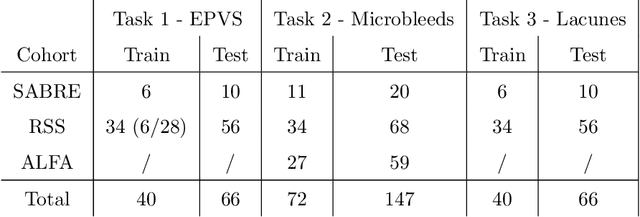
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022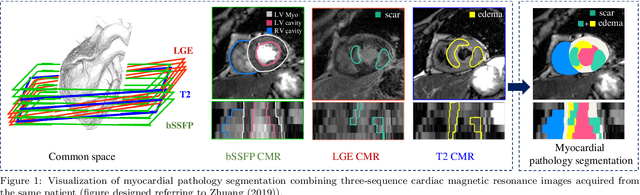
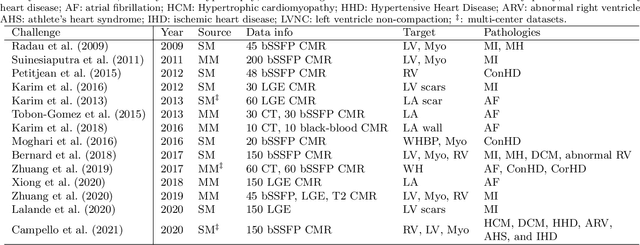
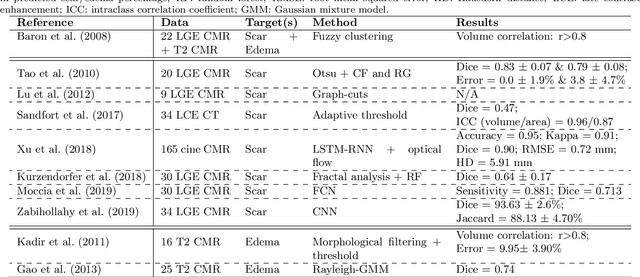
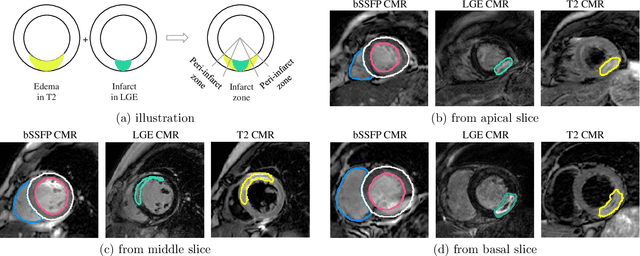
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021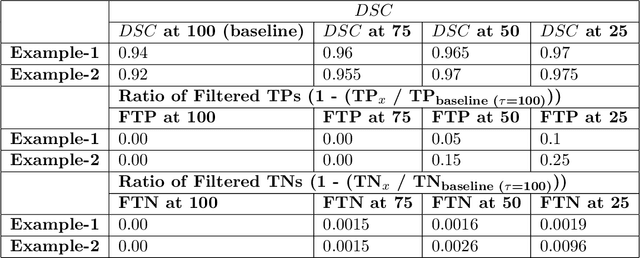
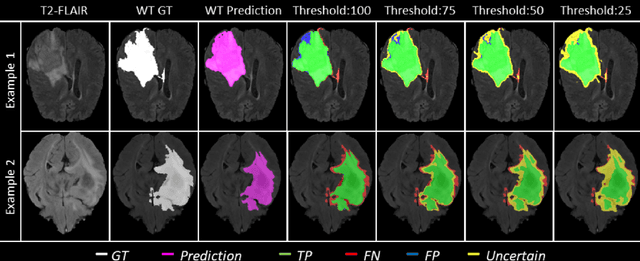
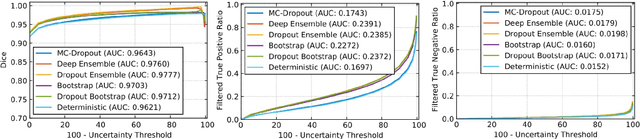
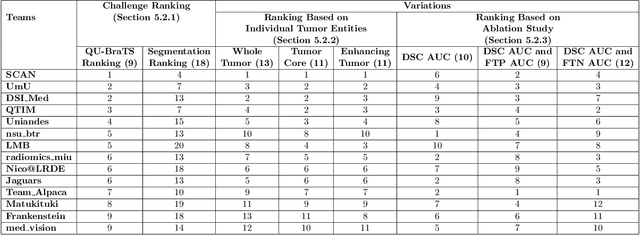
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Going beyond p-convolutions to learn grayscale morphological operators
Feb 19, 2021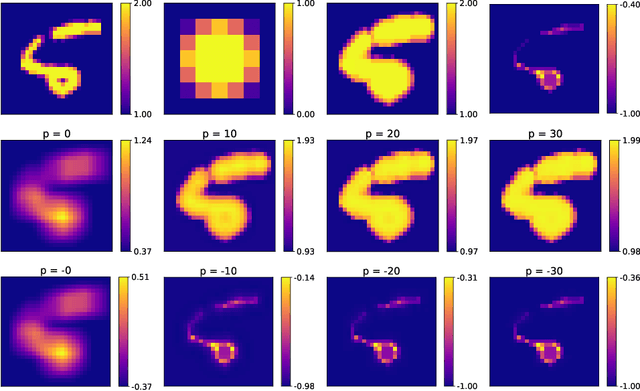
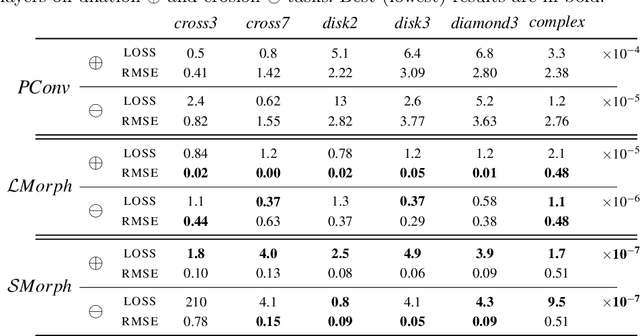
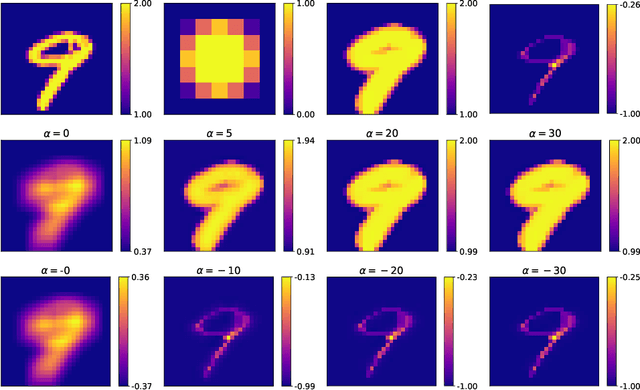
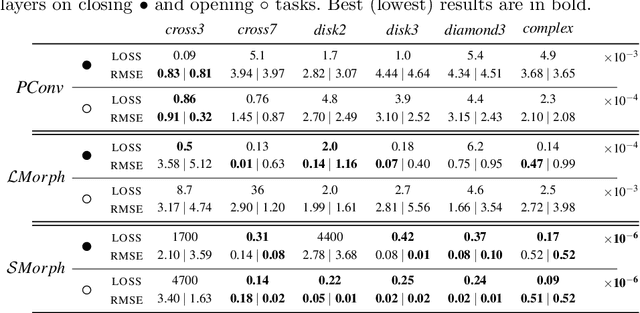
Abstract:Integrating mathematical morphology operations within deep neural networks has been subject to increasing attention lately. However, replacing standard convolution layers with erosions or dilations is particularly challenging because the min and max operations are not differentiable. Relying on the asymptotic behavior of the counter-harmonic mean, p-convolutional layers were proposed as a possible workaround to this issue since they can perform pseudo-dilation or pseudo-erosion operations (depending on the value of their inner parameter p), and very promising results were reported. In this work, we present two new morphological layers based on the same principle as the p-convolutional layer while circumventing its principal drawbacks, and demonstrate their potential interest in further implementations within deep convolutional neural network architectures.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020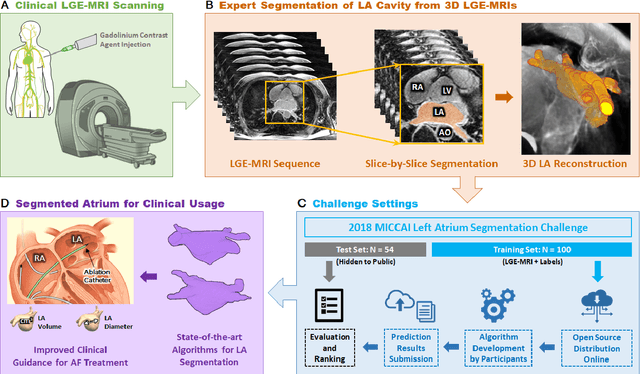
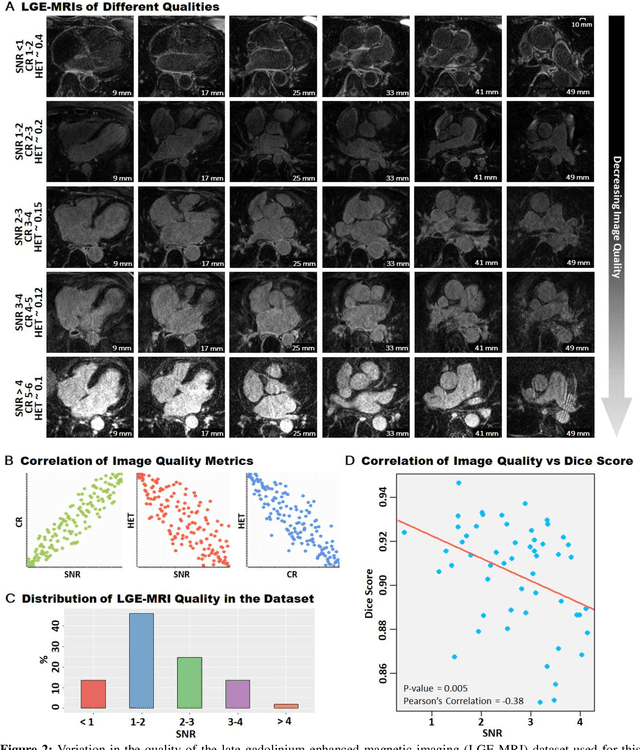
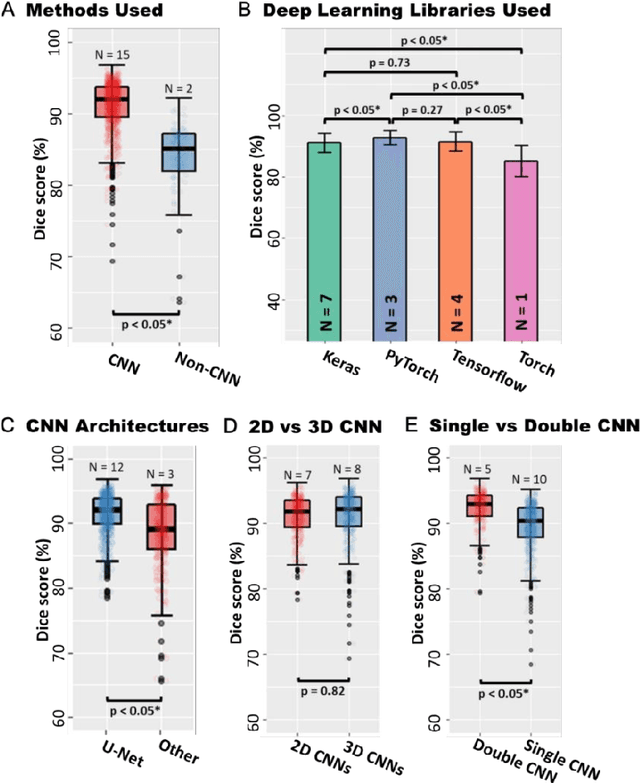
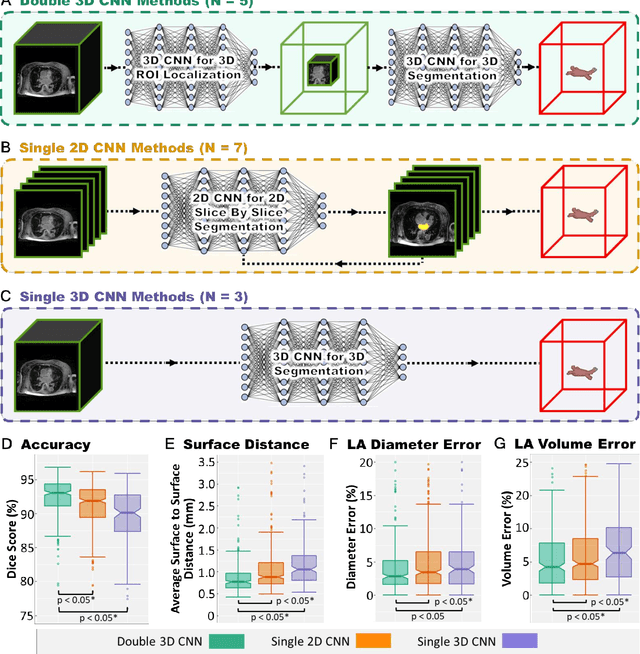
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Standardized Assessment of Automatic Segmentation of White Matter Hyperintensities and Results of the WMH Segmentation Challenge
Apr 01, 2019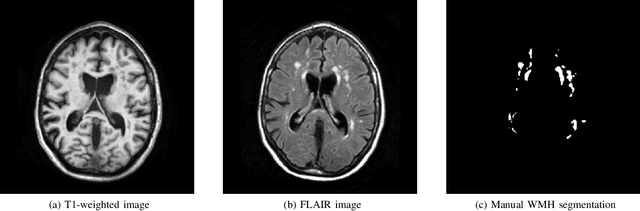
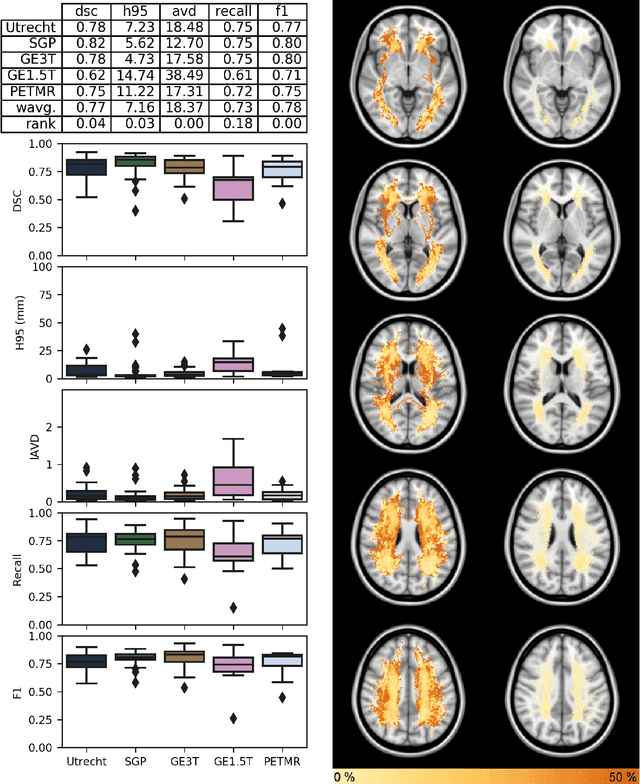
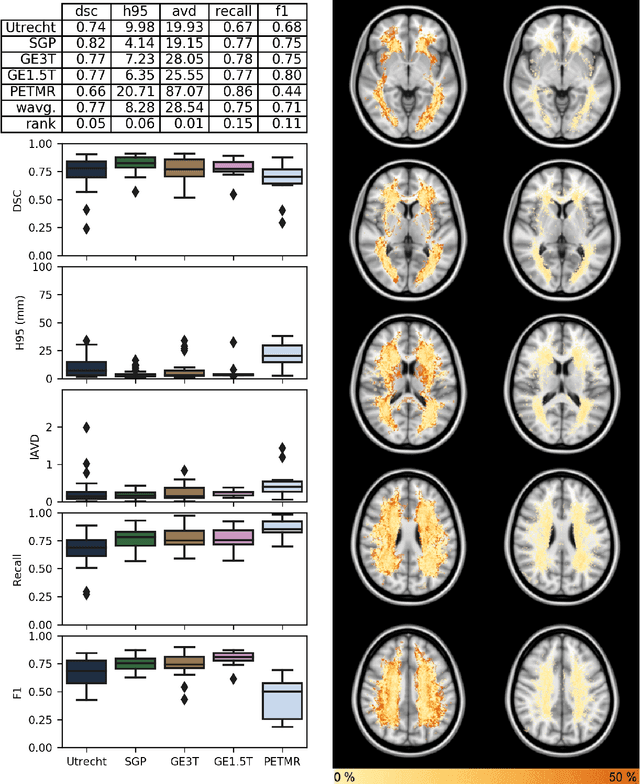
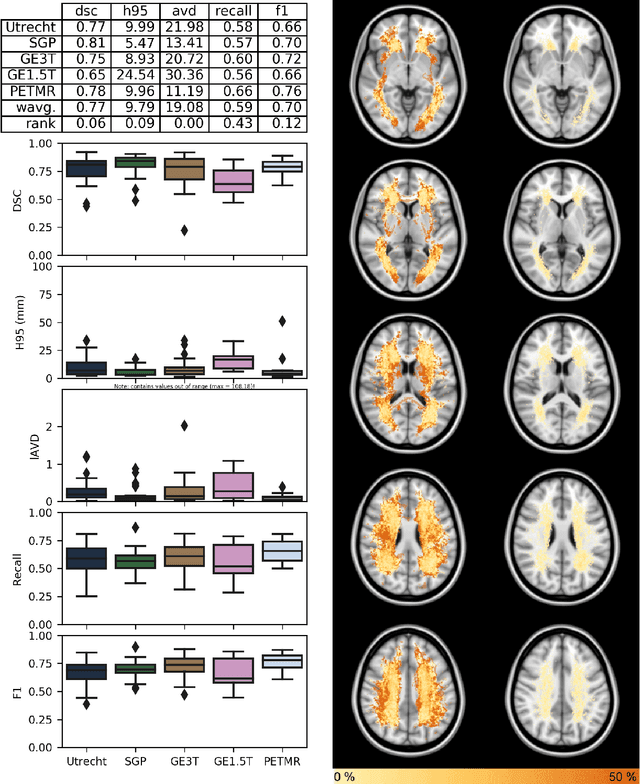
Abstract:Quantification of cerebral white matter hyperintensities (WMH) of presumed vascular origin is of key importance in many neurological research studies. Currently, measurements are often still obtained from manual segmentations on brain MR images, which is a laborious procedure. Automatic WMH segmentation methods exist, but a standardized comparison of the performance of such methods is lacking. We organized a scientific challenge, in which developers could evaluate their method on a standardized multi-center/-scanner image dataset, giving an objective comparison: the WMH Segmentation Challenge (https://wmh.isi.uu.nl/). Sixty T1+FLAIR images from three MR scanners were released with manual WMH segmentations for training. A test set of 110 images from five MR scanners was used for evaluation. Segmentation methods had to be containerized and submitted to the challenge organizers. Five evaluation metrics were used to rank the methods: (1) Dice similarity coefficient, (2) modified Hausdorff distance (95th percentile), (3) absolute log-transformed volume difference, (4) sensitivity for detecting individual lesions, and (5) F1-score for individual lesions. Additionally, methods were ranked on their inter-scanner robustness. Twenty participants submitted their method for evaluation. This paper provides a detailed analysis of the results. In brief, there is a cluster of four methods that rank significantly better than the other methods, with one clear winner. The inter-scanner robustness ranking shows that not all methods generalize to unseen scanners. The challenge remains open for future submissions and provides a public platform for method evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge