Guillaume Tochon
GIPSA-SIGMAPHY
Augmented Invertible Koopman Autoencoder for long-term time series forecasting
Mar 17, 2025Abstract:Following the introduction of Dynamic Mode Decomposition and its numerous extensions, many neural autoencoder-based implementations of the Koopman operator have recently been proposed. This class of methods appears to be of interest for modeling dynamical systems, either through direct long-term prediction of the evolution of the state or as a powerful embedding for downstream methods. In particular, a recent line of work has developed invertible Koopman autoencoders (IKAEs), which provide an exact reconstruction of the input state thanks to their analytically invertible encoder, based on coupling layer normalizing flow models. We identify that the conservation of the dimension imposed by the normalizing flows is a limitation for the IKAE models, and thus we propose to augment the latent state with a second, non-invertible encoder network. This results in our new model: the Augmented Invertible Koopman AutoEncoder (AIKAE). We demonstrate the relevance of the AIKAE through a series of long-term time series forecasting experiments, on satellite image time series as well as on a benchmark involving predictions based on a large lookback window of observations.
Koopman Ensembles for Probabilistic Time Series Forecasting
Mar 13, 2024


Abstract:In the context of an increasing popularity of data-driven models to represent dynamical systems, many machine learning-based implementations of the Koopman operator have recently been proposed. However, the vast majority of those works are limited to deterministic predictions, while the knowledge of uncertainty is critical in fields like meteorology and climatology. In this work, we investigate the training of ensembles of models to produce stochastic outputs. We show through experiments on real remote sensing image time series that ensembles of independently trained models are highly overconfident and that using a training criterion that explicitly encourages the members to produce predictions with high inter-model variances greatly improves the uncertainty quantification of the ensembles.
Neural Koopman prior for data assimilation
Sep 11, 2023Abstract:With the increasing availability of large scale datasets, computational power and tools like automatic differentiation and expressive neural network architectures, sequential data are now often treated in a data-driven way, with a dynamical model trained from the observation data. While neural networks are often seen as uninterpretable black-box architectures, they can still benefit from physical priors on the data and from mathematical knowledge. In this paper, we use a neural network architecture which leverages the long-known Koopman operator theory to embed dynamical systems in latent spaces where their dynamics can be described linearly, enabling a number of appealing features. We introduce methods that enable to train such a model for long-term continuous reconstruction, even in difficult contexts where the data comes in irregularly-sampled time series. The potential for self-supervised learning is also demonstrated, as we show the promising use of trained dynamical models as priors for variational data assimilation techniques, with applications to e.g. time series interpolation and forecasting.
Learning Sentinel-2 reflectance dynamics for data-driven assimilation and forecasting
May 05, 2023Abstract:Over the last few years, massive amounts of satellite multispectral and hyperspectral images covering the Earth's surface have been made publicly available for scientific purpose, for example through the European Copernicus project. Simultaneously, the development of self-supervised learning (SSL) methods has sparked great interest in the remote sensing community, enabling to learn latent representations from unlabeled data to help treating downstream tasks for which there is few annotated examples, such as interpolation, forecasting or unmixing. Following this line, we train a deep learning model inspired from the Koopman operator theory to model long-term reflectance dynamics in an unsupervised way. We show that this trained model, being differentiable, can be used as a prior for data assimilation in a straightforward way. Our datasets, which are composed of Sentinel-2 multispectral image time series, are publicly released with several levels of treatment.
Why is the winner the best?
Mar 30, 2023



Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Leveraging Neural Koopman Operators to Learn Continuous Representations of Dynamical Systems from Scarce Data
Mar 13, 2023Abstract:Over the last few years, several works have proposed deep learning architectures to learn dynamical systems from observation data with no or little knowledge of the underlying physics. A line of work relies on learning representations where the dynamics of the underlying phenomenon can be described by a linear operator, based on the Koopman operator theory. However, despite being able to provide reliable long-term predictions for some dynamical systems in ideal situations, the methods proposed so far have limitations, such as requiring to discretize intrinsically continuous dynamical systems, leading to data loss, especially when handling incomplete or sparsely sampled data. Here, we propose a new deep Koopman framework that represents dynamics in an intrinsically continuous way, leading to better performance on limited training data, as exemplified on several datasets arising from dynamical systems.
Where is VALDO? VAscular Lesions Detection and segmentatiOn challenge at MICCAI 2021
Aug 15, 2022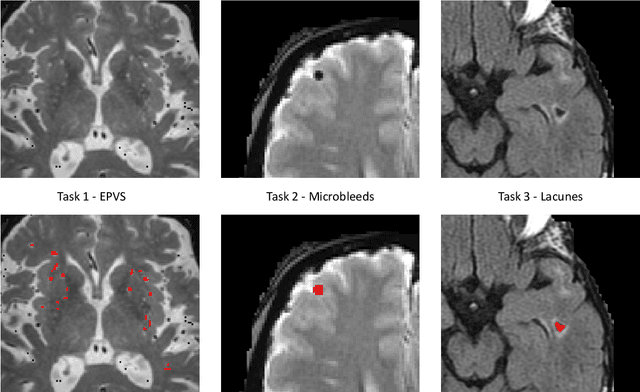
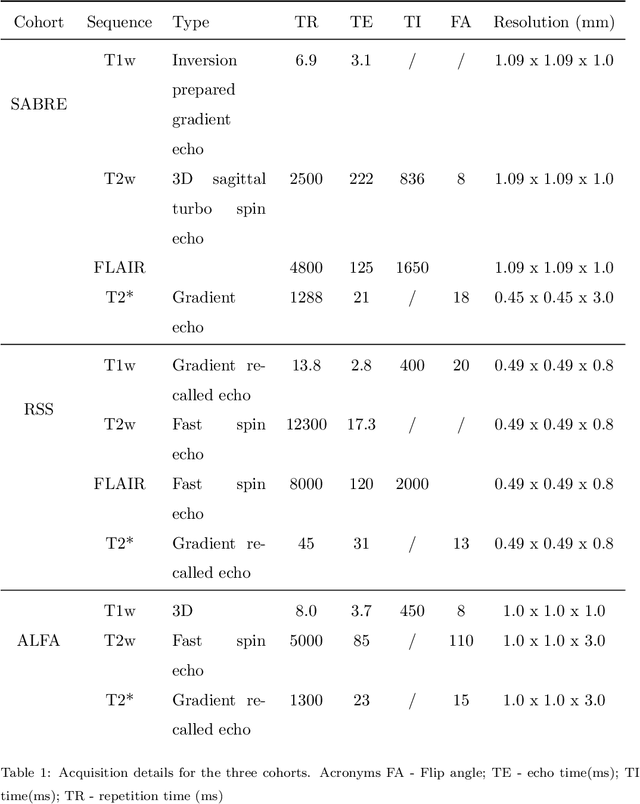
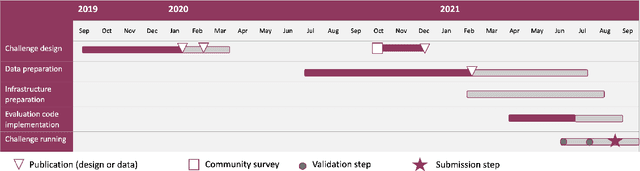
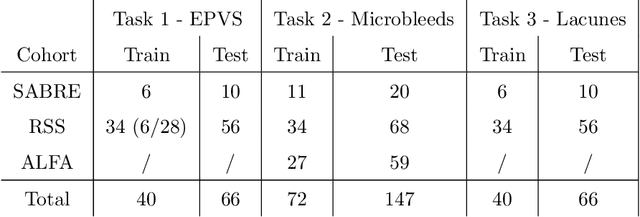
Abstract:Imaging markers of cerebral small vessel disease provide valuable information on brain health, but their manual assessment is time-consuming and hampered by substantial intra- and interrater variability. Automated rating may benefit biomedical research, as well as clinical assessment, but diagnostic reliability of existing algorithms is unknown. Here, we present the results of the \textit{VAscular Lesions DetectiOn and Segmentation} (\textit{Where is VALDO?}) challenge that was run as a satellite event at the international conference on Medical Image Computing and Computer Aided Intervention (MICCAI) 2021. This challenge aimed to promote the development of methods for automated detection and segmentation of small and sparse imaging markers of cerebral small vessel disease, namely enlarged perivascular spaces (EPVS) (Task 1), cerebral microbleeds (Task 2) and lacunes of presumed vascular origin (Task 3) while leveraging weak and noisy labels. Overall, 12 teams participated in the challenge proposing solutions for one or more tasks (4 for Task 1 - EPVS, 9 for Task 2 - Microbleeds and 6 for Task 3 - Lacunes). Multi-cohort data was used in both training and evaluation. Results showed a large variability in performance both across teams and across tasks, with promising results notably for Task 1 - EPVS and Task 2 - Microbleeds and not practically useful results yet for Task 3 - Lacunes. It also highlighted the performance inconsistency across cases that may deter use at an individual level, while still proving useful at a population level.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021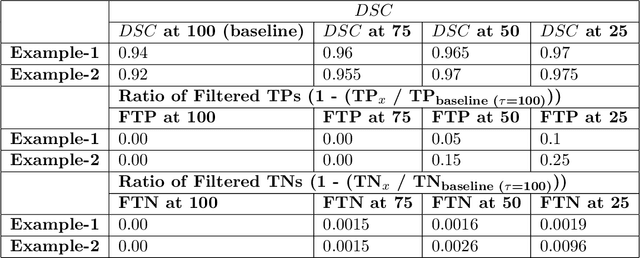
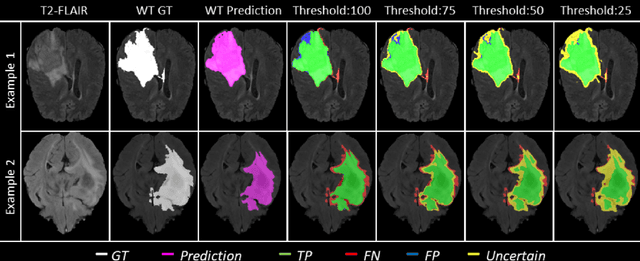
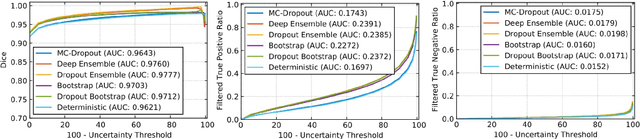
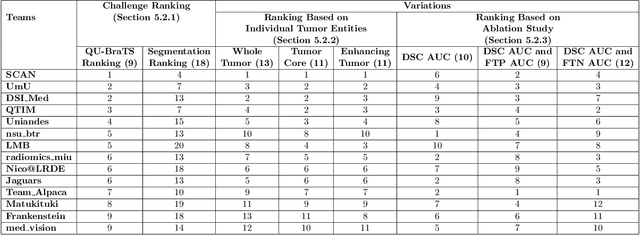
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Going beyond p-convolutions to learn grayscale morphological operators
Feb 19, 2021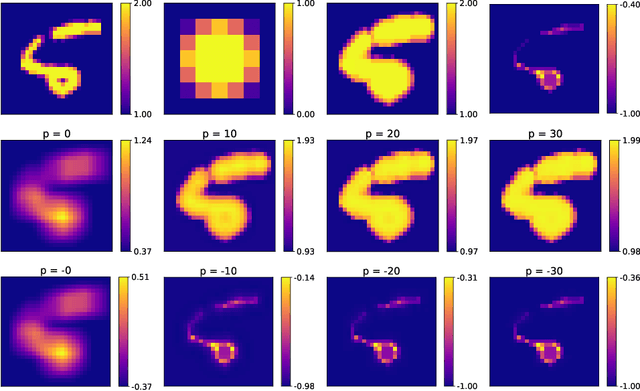
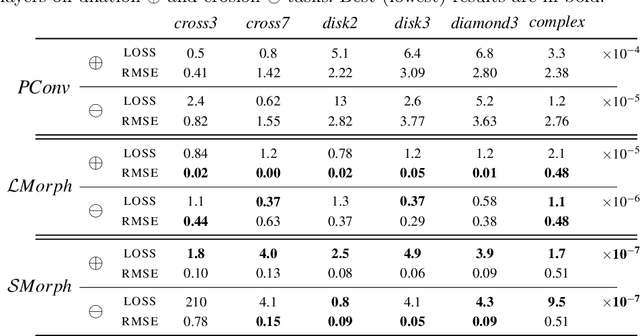
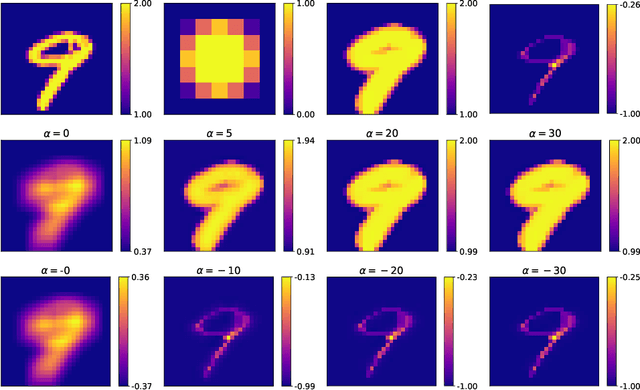
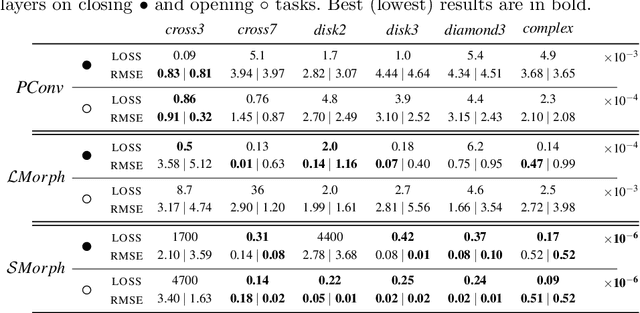
Abstract:Integrating mathematical morphology operations within deep neural networks has been subject to increasing attention lately. However, replacing standard convolution layers with erosions or dilations is particularly challenging because the min and max operations are not differentiable. Relying on the asymptotic behavior of the counter-harmonic mean, p-convolutional layers were proposed as a possible workaround to this issue since they can perform pseudo-dilation or pseudo-erosion operations (depending on the value of their inner parameter p), and very promising results were reported. In this work, we present two new morphological layers based on the same principle as the p-convolutional layer while circumventing its principal drawbacks, and demonstrate their potential interest in further implementations within deep convolutional neural network architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge