Apaar Sadhwani
AugLift: Boosting Generalization in Lifting-based 3D Human Pose Estimation
Aug 09, 2025



Abstract:Lifting-based methods for 3D Human Pose Estimation (HPE), which predict 3D poses from detected 2D keypoints, often generalize poorly to new datasets and real-world settings. To address this, we propose \emph{AugLift}, a simple yet effective reformulation of the standard lifting pipeline that significantly improves generalization performance without requiring additional data collection or sensors. AugLift sparsely enriches the standard input -- the 2D keypoint coordinates $(x, y)$ -- by augmenting it with a keypoint detection confidence score $c$ and a corresponding depth estimate $d$. These additional signals are computed from the image using off-the-shelf, pre-trained models (e.g., for monocular depth estimation), thereby inheriting their strong generalization capabilities. Importantly, AugLift serves as a modular add-on and can be readily integrated into existing lifting architectures. Our extensive experiments across four datasets demonstrate that AugLift boosts cross-dataset performance on unseen datasets by an average of $10.1\%$, while also improving in-distribution performance by $4.0\%$. These gains are consistent across various lifting architectures, highlighting the robustness of our method. Our analysis suggests that these sparse, keypoint-aligned cues provide robust frame-level context, offering a practical way to significantly improve the generalization of any lifting-based pose estimation model. Code will be made publicly available.
A Set-Sequence Model for Time Series
May 16, 2025Abstract:In many financial prediction problems, the behavior of individual units (such as loans, bonds, or stocks) is influenced by observable unit-level factors and macroeconomic variables, as well as by latent cross-sectional effects. Traditional approaches attempt to capture these latent effects via handcrafted summary features. We propose a Set-Sequence model that eliminates the need for handcrafted features. The Set model first learns a shared cross-sectional summary at each period. The Sequence model then ingests the summary-augmented time series for each unit independently to predict its outcome. Both components are learned jointly over arbitrary sets sampled during training. Our approach harnesses the set nature of the cross-section and is computationally efficient, generating set summaries in linear time relative to the number of units. It is also flexible, allowing the use of existing sequence models and accommodating a variable number of units at inference. Empirical evaluations demonstrate that our Set-Sequence model significantly outperforms benchmarks on stock return prediction and mortgage behavior tasks. Code will be released.
Interpretable Survival Prediction for Colorectal Cancer using Deep Learning
Nov 17, 2020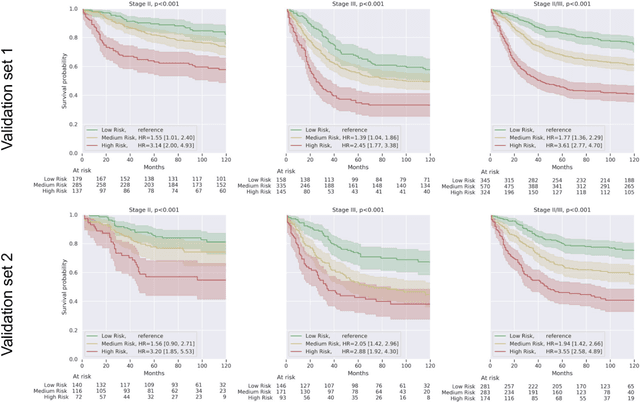
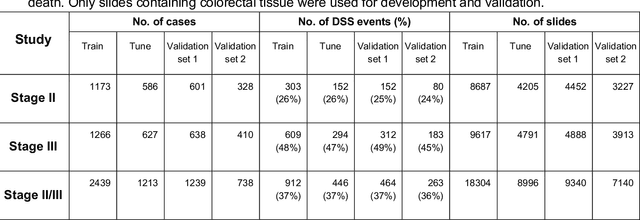
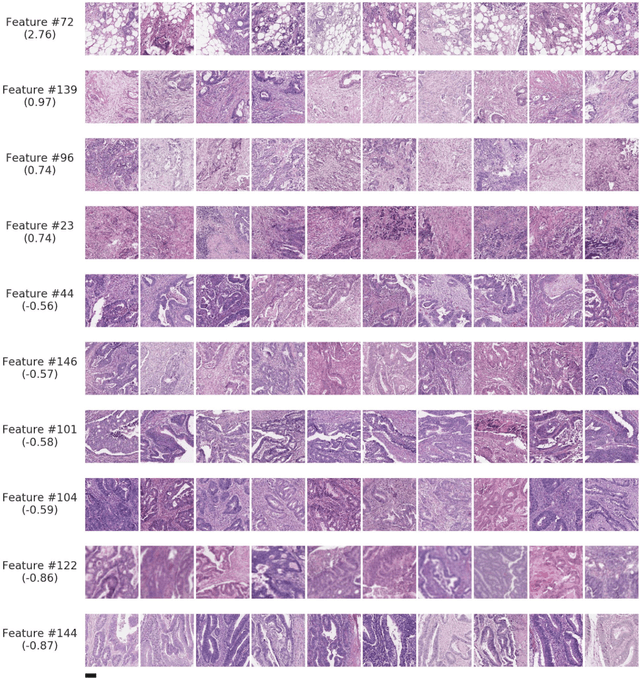
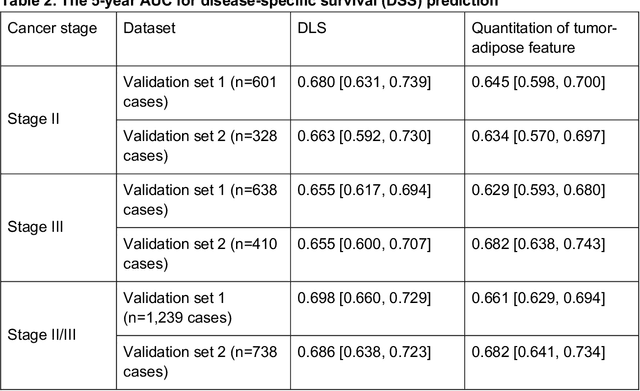
Abstract:Deriving interpretable prognostic features from deep-learning-based prognostic histopathology models remains a challenge. In this study, we developed a deep learning system (DLS) for predicting disease specific survival for stage II and III colorectal cancer using 3,652 cases (27,300 slides). When evaluated on two validation datasets containing 1,239 cases (9,340 slides) and 738 cases (7,140 slides) respectively, the DLS achieved a 5-year disease-specific survival AUC of 0.70 (95%CI 0.66-0.73) and 0.69 (95%CI 0.64-0.72), and added significant predictive value to a set of 9 clinicopathologic features. To interpret the DLS, we explored the ability of different human-interpretable features to explain the variance in DLS scores. We observed that clinicopathologic features such as T-category, N-category, and grade explained a small fraction of the variance in DLS scores (R2=18% in both validation sets). Next, we generated human-interpretable histologic features by clustering embeddings from a deep-learning based image-similarity model and showed that they explain the majority of the variance (R2 of 73% to 80%). Furthermore, the clustering-derived feature most strongly associated with high DLS scores was also highly prognostic in isolation. With a distinct visual appearance (poorly differentiated tumor cell clusters adjacent to adipose tissue), this feature was identified by annotators with 87.0-95.5% accuracy. Our approach can be used to explain predictions from a prognostic deep learning model and uncover potentially-novel prognostic features that can be reliably identified by people for future validation studies.
Deep learning-based survival prediction for multiple cancer types using histopathology images
Dec 16, 2019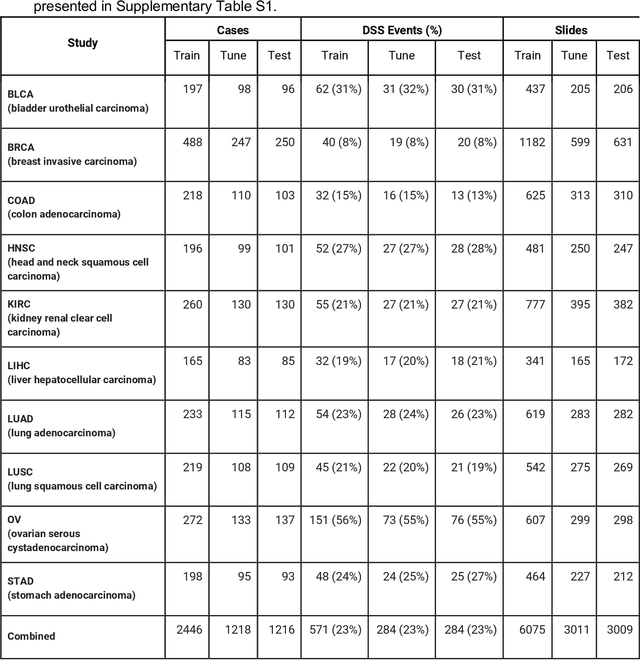
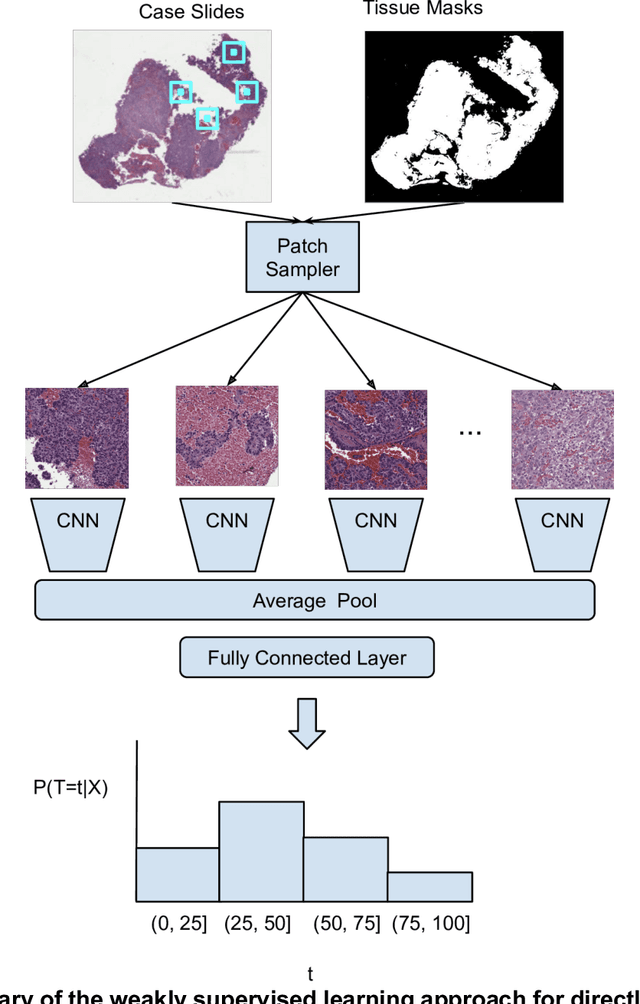
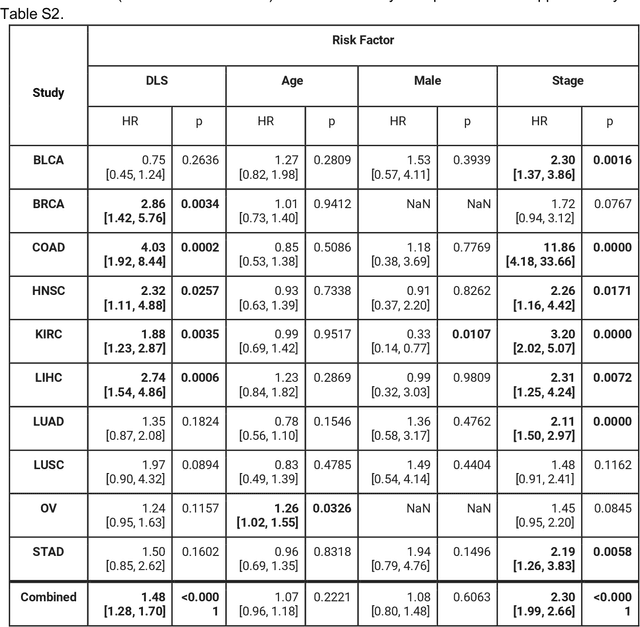
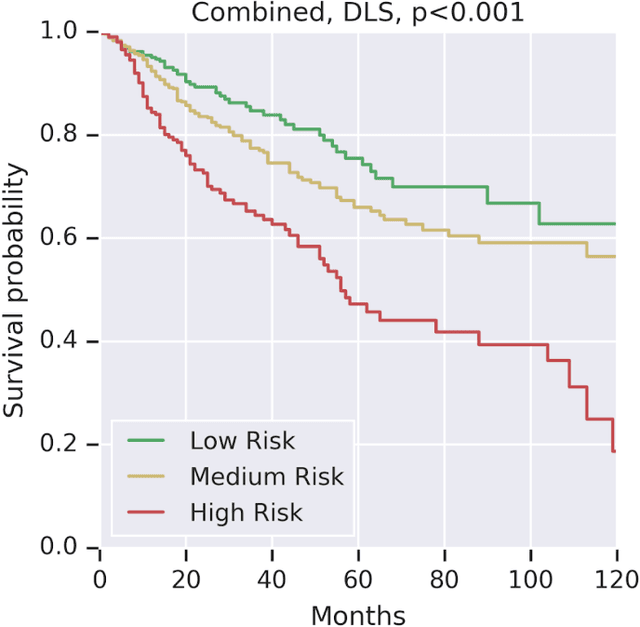
Abstract:Prognostic information at diagnosis has important implications for cancer treatment and monitoring. Although cancer staging, histopathological assessment, molecular features, and clinical variables can provide useful prognostic insights, improving risk stratification remains an active research area. We developed a deep learning system (DLS) to predict disease specific survival across 10 cancer types from The Cancer Genome Atlas (TCGA). We used a weakly-supervised approach without pixel-level annotations, and tested three different survival loss functions. The DLS was developed using 9,086 slides from 3,664 cases and evaluated using 3,009 slides from 1,216 cases. In multivariable Cox regression analysis of the combined cohort including all 10 cancers, the DLS was significantly associated with disease specific survival (hazard ratio of 1.58, 95% CI 1.28-1.70, p<0.0001) after adjusting for cancer type, stage, age, and sex. In a per-cancer adjusted subanalysis, the DLS remained a significant predictor of survival in 5 of 10 cancer types. Compared to a baseline model including stage, age, and sex, the c-index of the model demonstrated an absolute 3.7% improvement (95% CI 1.0-6.5) in the combined cohort. Additionally, our models stratified patients within individual cancer stages, particularly stage II (p=0.025) and stage III (p<0.001). By developing and evaluating prognostic models across multiple cancer types, this work represents one of the most comprehensive studies exploring the direct prediction of clinical outcomes using deep learning and histopathology images. Our analysis demonstrates the potential for this approach to provide prognostic information in multiple cancer types, and even within specific pathologic stages. However, given the relatively small number of clinical events, we observed wide confidence intervals, suggesting that future work will benefit from larger datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge