Sebastien Baur
Health AI Developer Foundations
Nov 26, 2024



Abstract:Robust medical Machine Learning (ML) models have the potential to revolutionize healthcare by accelerating clinical research, improving workflows and outcomes, and producing novel insights or capabilities. Developing such ML models from scratch is cost prohibitive and requires substantial compute, data, and time (e.g., expert labeling). To address these challenges, we introduce Health AI Developer Foundations (HAI-DEF), a suite of pre-trained, domain-specific foundation models, tools, and recipes to accelerate building ML for health applications. The models cover various modalities and domains, including radiology (X-rays and computed tomography), histopathology, dermatological imaging, and audio. These models provide domain specific embeddings that facilitate AI development with less labeled data, shorter training times, and reduced computational costs compared to traditional approaches. In addition, we utilize a common interface and style across these models, and prioritize usability to enable developers to integrate HAI-DEF efficiently. We present model evaluations across various tasks and conclude with a discussion of their application and evaluation, covering the importance of ensuring efficacy, fairness, and equity. Finally, while HAI-DEF and specifically the foundation models lower the barrier to entry for ML in healthcare, we emphasize the importance of validation with problem- and population-specific data for each desired usage setting. This technical report will be updated over time as more modalities and features are added.
HeAR -- Health Acoustic Representations
Mar 04, 2024



Abstract:Health acoustic sounds such as coughs and breaths are known to contain useful health signals with significant potential for monitoring health and disease, yet are underexplored in the medical machine learning community. The existing deep learning systems for health acoustics are often narrowly trained and evaluated on a single task, which is limited by data and may hinder generalization to other tasks. To mitigate these gaps, we develop HeAR, a scalable self-supervised learning-based deep learning system using masked autoencoders trained on a large dataset of 313 million two-second long audio clips. Through linear probes, we establish HeAR as a state-of-the-art health audio embedding model on a benchmark of 33 health acoustic tasks across 6 datasets. By introducing this work, we hope to enable and accelerate further health acoustics research.
Optimizing Audio Augmentations for Contrastive Learning of Health-Related Acoustic Signals
Sep 11, 2023

Abstract:Health-related acoustic signals, such as cough and breathing sounds, are relevant for medical diagnosis and continuous health monitoring. Most existing machine learning approaches for health acoustics are trained and evaluated on specific tasks, limiting their generalizability across various healthcare applications. In this paper, we leverage a self-supervised learning framework, SimCLR with a Slowfast NFNet backbone, for contrastive learning of health acoustics. A crucial aspect of optimizing Slowfast NFNet for this application lies in identifying effective audio augmentations. We conduct an in-depth analysis of various audio augmentation strategies and demonstrate that an appropriate augmentation strategy enhances the performance of the Slowfast NFNet audio encoder across a diverse set of health acoustic tasks. Our findings reveal that when augmentations are combined, they can produce synergistic effects that exceed the benefits seen when each is applied individually.
Predicting Cardiovascular Disease Risk using Photoplethysmography and Deep Learning
May 09, 2023Abstract:Cardiovascular diseases (CVDs) are responsible for a large proportion of premature deaths in low- and middle-income countries. Early CVD detection and intervention is critical in these populations, yet many existing CVD risk scores require a physical examination or lab measurements, which can be challenging in such health systems due to limited accessibility. Here we investigated the potential to use photoplethysmography (PPG), a sensing technology available on most smartphones that can potentially enable large-scale screening at low cost, for CVD risk prediction. We developed a deep learning PPG-based CVD risk score (DLS) to predict the probability of having major adverse cardiovascular events (MACE: non-fatal myocardial infarction, stroke, and cardiovascular death) within ten years, given only age, sex, smoking status and PPG as predictors. We compared the DLS with the office-based refit-WHO score, which adopts the shared predictors from WHO and Globorisk scores (age, sex, smoking status, height, weight and systolic blood pressure) but refitted on the UK Biobank (UKB) cohort. In UKB cohort, DLS's C-statistic (71.1%, 95% CI 69.9-72.4) was non-inferior to office-based refit-WHO score (70.9%, 95% CI 69.7-72.2; non-inferiority margin of 2.5%, p<0.01). The calibration of the DLS was satisfactory, with a 1.8% mean absolute calibration error. Adding DLS features to the office-based score increased the C-statistic by 1.0% (95% CI 0.6-1.4). DLS predicts ten-year MACE risk comparable with the office-based refit-WHO score. It provides a proof-of-concept and suggests the potential of a PPG-based approach strategies for community-based primary prevention in resource-limited regions.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022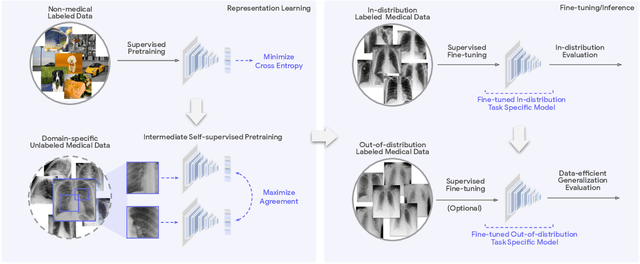
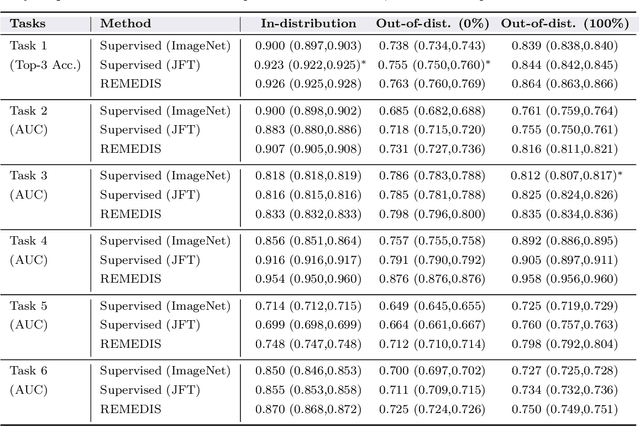
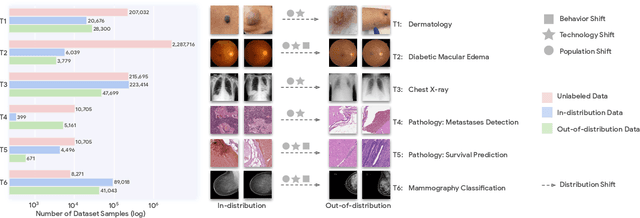
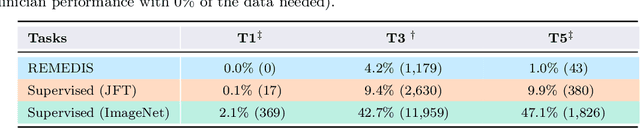
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
Best of both worlds: local and global explanations with human-understandable concepts
Jun 16, 2021



Abstract:Interpretability techniques aim to provide the rationale behind a model's decision, typically by explaining either an individual prediction (local explanation, e.g. `why is this patient diagnosed with this condition') or a class of predictions (global explanation, e.g. `why are patients diagnosed with this condition in general'). While there are many methods focused on either one, few frameworks can provide both local and global explanations in a consistent manner. In this work, we combine two powerful existing techniques, one local (Integrated Gradients, IG) and one global (Testing with Concept Activation Vectors), to provide local, and global concept-based explanations. We first validate our idea using two synthetic datasets with a known ground truth, and further demonstrate with a benchmark natural image dataset. We test our method with various concepts, target classes, model architectures and IG baselines. We show that our method improves global explanations over TCAV when compared to ground truth, and provides useful insights. We hope our work provides a step towards building bridges between many existing local and global methods to get the best of both worlds.
Concept-based model explanations for Electronic Health Records
Dec 03, 2020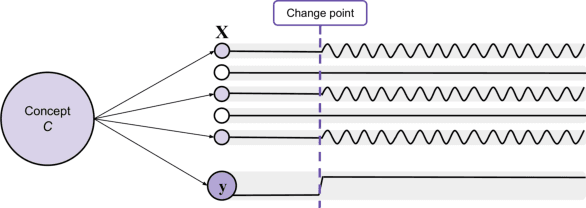
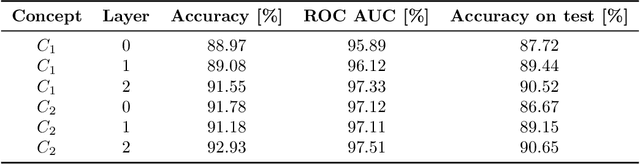
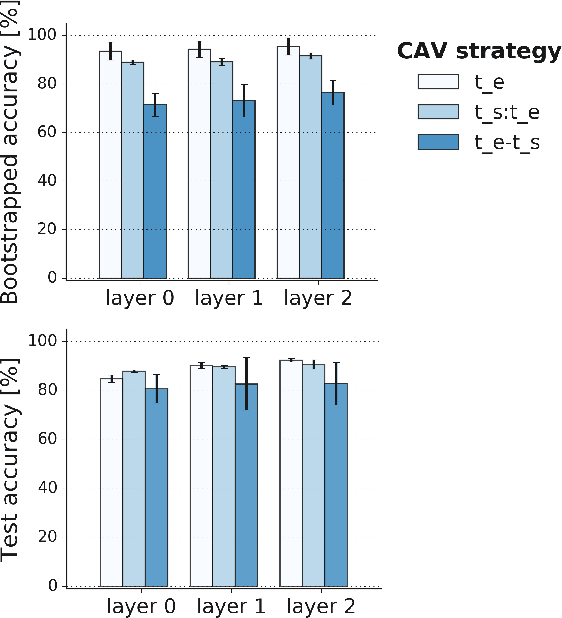
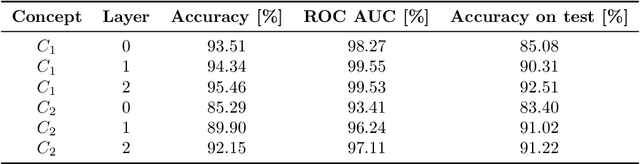
Abstract:Recurrent Neural Networks (RNNs) are often used for sequential modeling of adverse outcomes in electronic health records (EHRs) due to their ability to encode past clinical states. These deep, recurrent architectures have displayed increased performance compared to other modeling approaches in a number of tasks, fueling the interest in deploying deep models in clinical settings. One of the key elements in ensuring safe model deployment and building user trust is model explainability. Testing with Concept Activation Vectors (TCAV) has recently been introduced as a way of providing human-understandable explanations by comparing high-level concepts to the network's gradients. While the technique has shown promising results in real-world imaging applications, it has not been applied to structured temporal inputs. To enable an application of TCAV to sequential predictions in the EHR, we propose an extension of the method to time series data. We evaluate the proposed approach on an open EHR benchmark from the intensive care unit, as well as synthetic data where we are able to better isolate individual effects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge