Daniel Rubin
Advancing COVID-19 Diagnosis with Privacy-Preserving Collaboration in Artificial Intelligence
Nov 18, 2021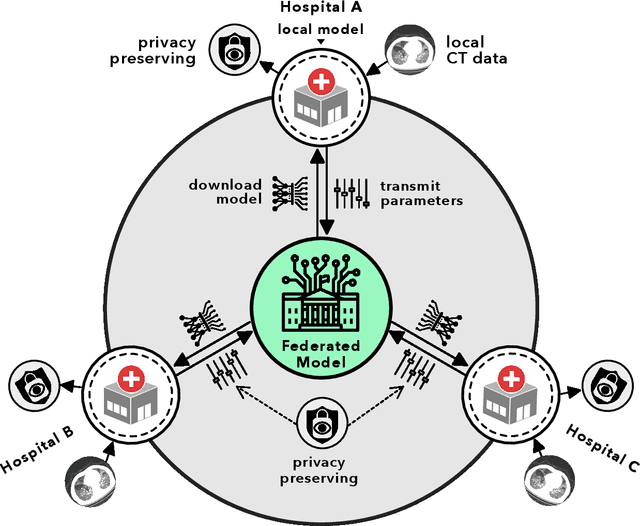
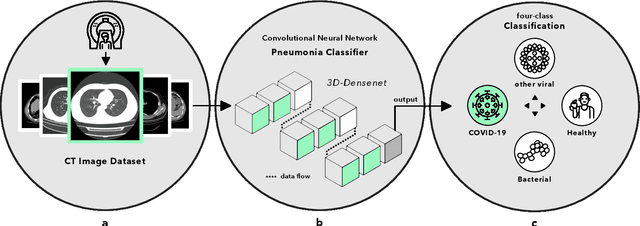
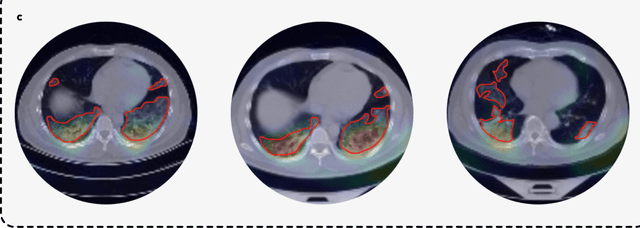
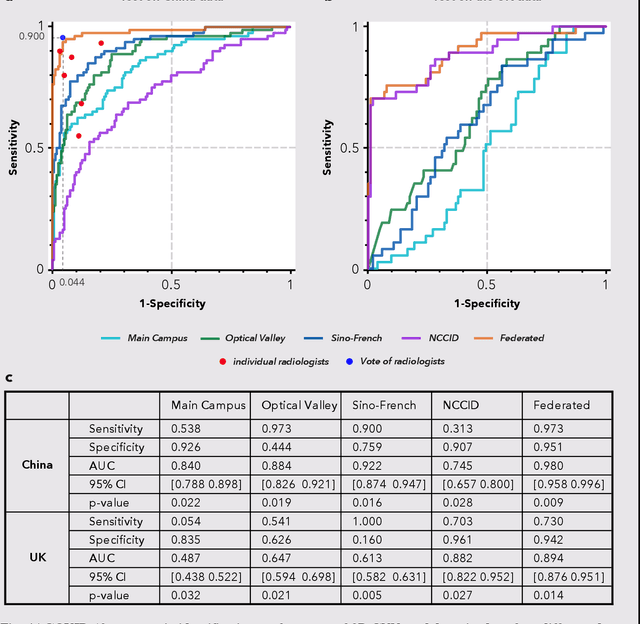
Abstract:Artificial intelligence (AI) provides a promising substitution for streamlining COVID-19 diagnoses. However, concerns surrounding security and trustworthiness impede the collection of large-scale representative medical data, posing a considerable challenge for training a well-generalised model in clinical practices. To address this, we launch the Unified CT-COVID AI Diagnostic Initiative (UCADI), where the AI model can be distributedly trained and independently executed at each host institution under a federated learning framework (FL) without data sharing. Here we show that our FL model outperformed all the local models by a large yield (test sensitivity /specificity in China: 0.973/0.951, in the UK: 0.730/0.942), achieving comparable performance with a panel of professional radiologists. We further evaluated the model on the hold-out (collected from another two hospitals leaving out the FL) and heterogeneous (acquired with contrast materials) data, provided visual explanations for decisions made by the model, and analysed the trade-offs between the model performance and the communication costs in the federated training process. Our study is based on 9,573 chest computed tomography scans (CTs) from 3,336 patients collected from 23 hospitals located in China and the UK. Collectively, our work advanced the prospects of utilising federated learning for privacy-preserving AI in digital health.
Handling Data Heterogeneity with Generative Replay in Collaborative Learning for Medical Imaging
Jun 24, 2021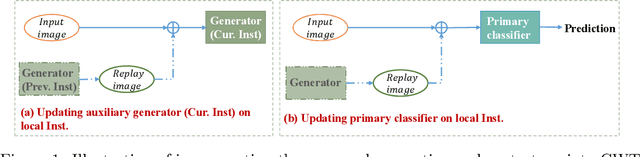
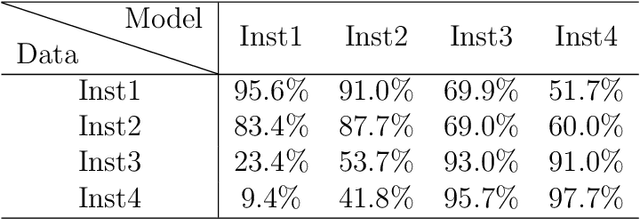
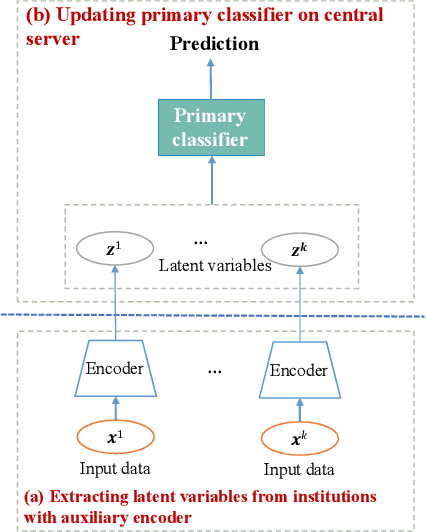
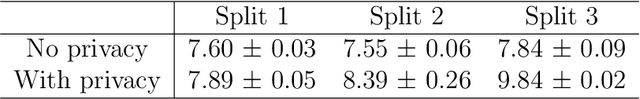
Abstract:Collaborative learning, which enables collaborative and decentralized training of deep neural networks at multiple institutions in a privacy-preserving manner, is rapidly emerging as a valuable technique in healthcare applications. However, its distributed nature often leads to significant heterogeneity in data distributions across institutions. Existing collaborative learning approaches generally do not account for the presence of heterogeneity in data among institutions, or only mildly skewed label distributions are studied. In this paper, we present a novel generative replay strategy to address the challenge of data heterogeneity in collaborative learning methods. Instead of directly training a model for task performance, we leverage recent image synthesis techniques to develop a novel dual model architecture: a primary model learns the desired task, and an auxiliary "generative replay model" either synthesizes images that closely resemble the input images or helps extract latent variables. The generative replay strategy is flexible to use, can either be incorporated into existing collaborative learning methods to improve their capability of handling data heterogeneity across institutions, or be used as a novel and individual collaborative learning framework (termed FedReplay) to reduce communication cost. Experimental results demonstrate the capability of the proposed method in handling heterogeneous data across institutions. On highly heterogeneous data partitions, our model achieves ~4.88% improvement in the prediction accuracy on a diabetic retinopathy classification dataset, and ~49.8% reduction of mean absolution value on a Bone Age prediction dataset, respectively, compared to the state-of-the art collaborative learning methods.
Rethinking Architecture Design for Tackling Data Heterogeneity in Federated Learning
Jun 10, 2021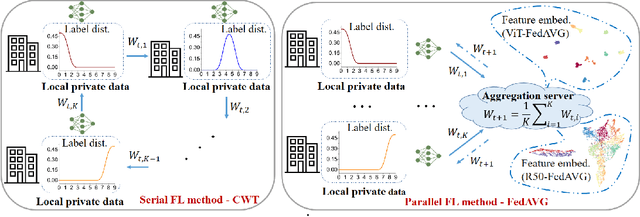
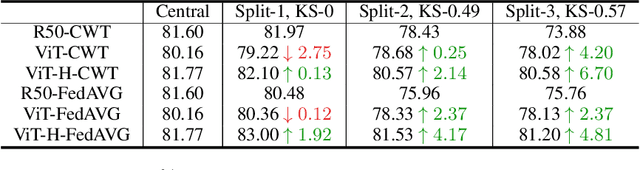
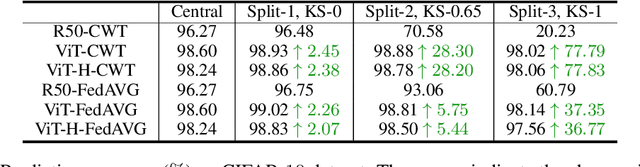
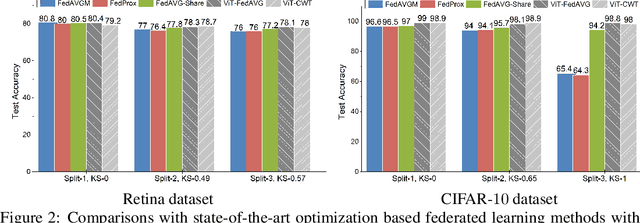
Abstract:Federated learning is an emerging research paradigm enabling collaborative training of machine learning models among different organizations while keeping data private at each institution. Despite recent progress, there remain fundamental challenges such as lack of convergence and potential for catastrophic forgetting in federated learning across real-world heterogeneous devices. In this paper, we demonstrate that attention-based architectures (e.g., Transformers) are fairly robust to distribution shifts and hence improve federated learning over heterogeneous data. Concretely, we conduct the first rigorous empirical investigation of different neural architectures across a range of federated algorithms, real-world benchmarks, and heterogeneous data splits. Our experiments show that simply replacing convolutional networks with Transformers can greatly reduce catastrophic forgetting of previous devices, accelerate convergence, and reach a better global model, especially when dealing with heterogeneous data. We will release our code and pretrained models at https://github.com/Liangqiong/ViT-FL-main to encourage future exploration in robust architectures as an alternative to current research efforts on the optimization front.
Double Descent Optimization Pattern and Aliasing: Caveats of Noisy Labels
Jun 03, 2021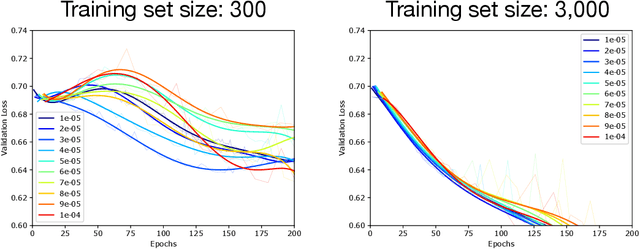
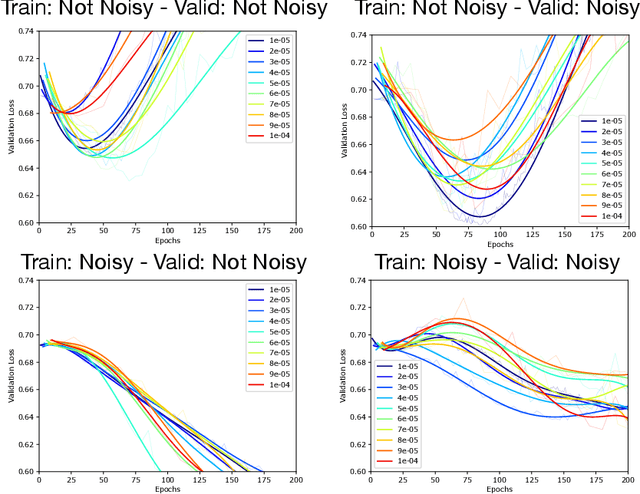
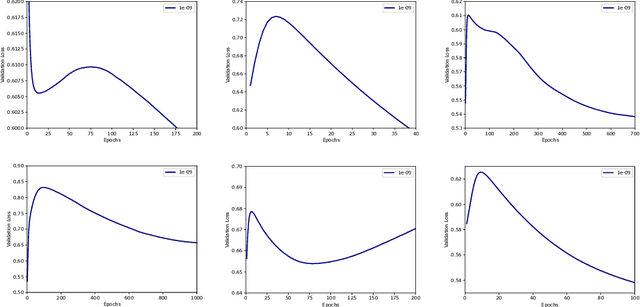
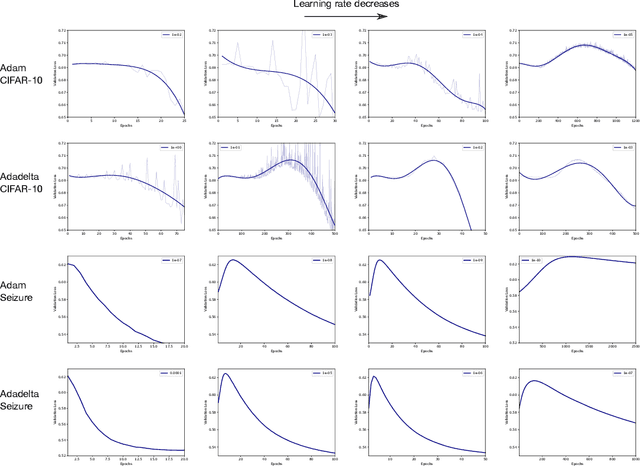
Abstract:Optimization plays a key role in the training of deep neural networks. Deciding when to stop training can have a substantial impact on the performance of the network during inference. Under certain conditions, the generalization error can display a double descent pattern during training: the learning curve is non-monotonic and seemingly diverges before converging again after additional epochs. This optimization pattern can lead to early stopping procedures to stop training before the second convergence and consequently select a suboptimal set of parameters for the network, with worse performance during inference. In this work, in addition to confirming that double descent occurs with small datasets and noisy labels as evidenced by others, we show that noisy labels must be present both in the training and generalization sets to observe a double descent pattern. We also show that the learning rate has an influence on double descent, and study how different optimizers and optimizer parameters influence the apparition of double descent. Finally, we show that increasing the learning rate can create an aliasing effect that masks the double descent pattern without suppressing it. We study this phenomenon through extensive experiments on variants of CIFAR-10 and show that they translate to a real world application: the forecast of seizure events in epileptic patients from continuous electroencephalographic recordings.
Inaccurate Supervision of Neural Networks with Incorrect Labels: Application to Epilepsy
Dec 01, 2020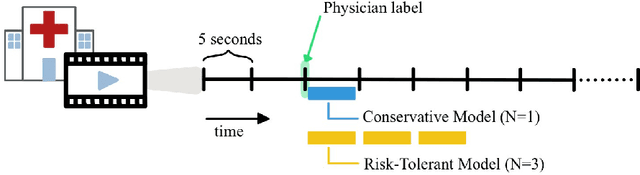
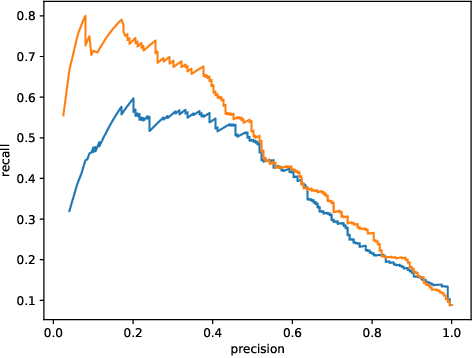
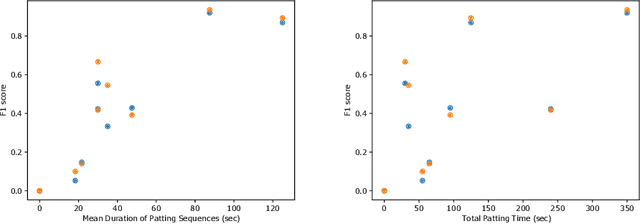
Abstract:This work describes multiple weak supervision strategies for video processing with neural networks in the context of seizure detection. To study seizure onset, we have designed automated methods to detect seizures from electroencephalography (EEG), a modality used for recording electrical brain activity. However, the EEG signal alone is sometimes not enough for existing detection methods to discriminate seizure from artifacts having a similar signal on EEG. For example, such artifacts could be triggered by patting, rocking or suctioning in the case of neonates. In this article, we addressed this problem by automatically detecting an example artifact -- patting of neonates -- from continuous video recordings of neonates acquired during clinical routine. We computed frame-to-frame cross-correlation matrices to isolate patterns showing repetitive movements indicative of patting of the patient. Next, a convolutional neural network was trained to classify whether these matrices contained patting events using weak training labels -- noisy labels generated during daily clinical procedure. The labels were considered weak as they were sometimes incorrect. We investigated whether networks trained with more samples, containing more uncertain and weak labels, could achieve a higher performance. Our results showed that, in the case of patting detection, such networks could achieve a higher recall, without sacrificing precision. These networks focused on areas of the cross-correlation matrices that were more meaningful to the task. More generally, our work gives insights into building more accurate models from weakly labelled time sequences.
Federated Learning for Breast Density Classification: A Real-World Implementation
Sep 17, 2020

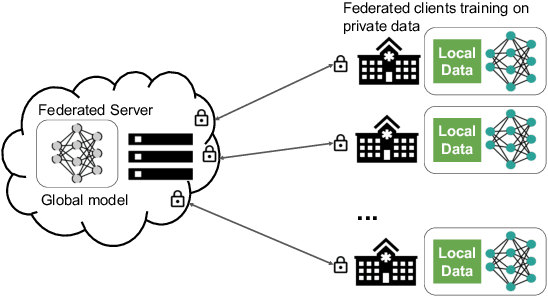
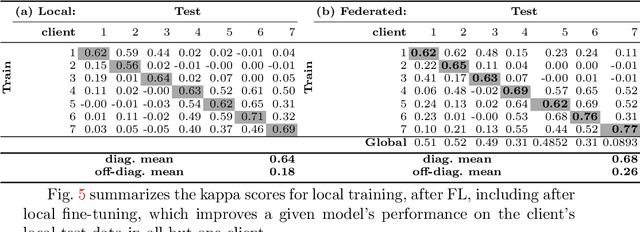
Abstract:Building robust deep learning-based models requires large quantities of diverse training data. In this study, we investigate the use of federated learning (FL) to build medical imaging classification models in a real-world collaborative setting. Seven clinical institutions from across the world joined this FL effort to train a model for breast density classification based on Breast Imaging, Reporting & Data System (BI-RADS). We show that despite substantial differences among the datasets from all sites (mammography system, class distribution, and data set size) and without centralizing data, we can successfully train AI models in federation. The results show that models trained using FL perform 6.3% on average better than their counterparts trained on an institute's local data alone. Furthermore, we show a 45.8% relative improvement in the models' generalizability when evaluated on the other participating sites' testing data.
Random Bundle: Brain Metastases Segmentation Ensembling through Annotation Randomization
Feb 23, 2020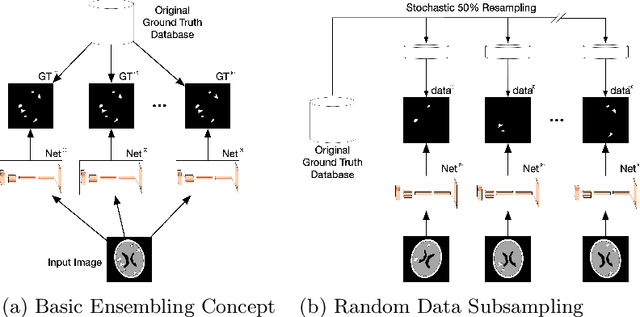
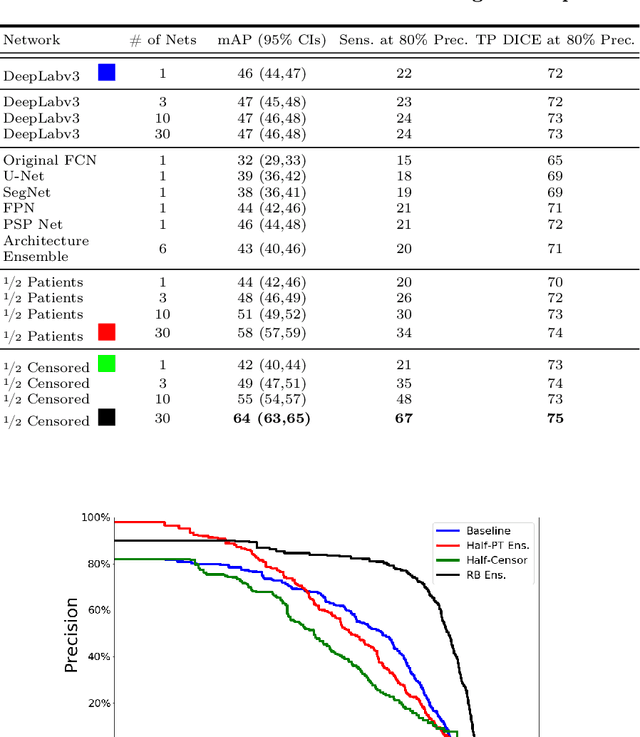
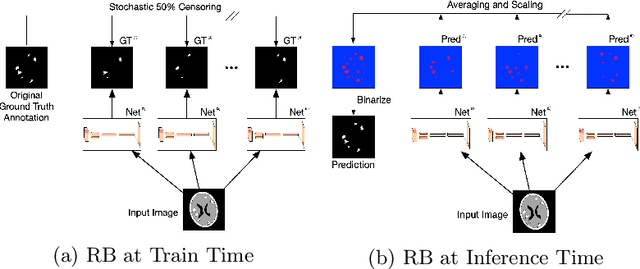
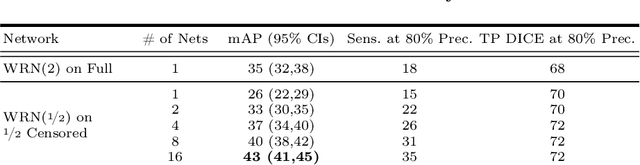
Abstract:We introduce a novel ensembling method, Random Bundle (RB), that improves performance for brain metastases segmentation. We create our ensemble by training each network on our dataset with 50% of our annotated lesions censored out. We also apply a lopsided bootstrap loss to recover performance after inducing an in silico 50% false negative rate and make our networks more sensitive. We improve our network detection of lesions's mAP value by 39% and more than triple the sensitivity at 80% precision. We also show slight improvements in segmentation quality through DICE score. Further, RB ensembling improves performance over baseline by a larger margin than a variety of popular ensembling strategies. Finally, we show that RB ensembling is computationally efficient by comparing its performance to a single network when both systems are constrained to have the same compute.
Brain Metastasis Segmentation Network Trained with Robustness to Annotations with Multiple False Negatives
Jan 26, 2020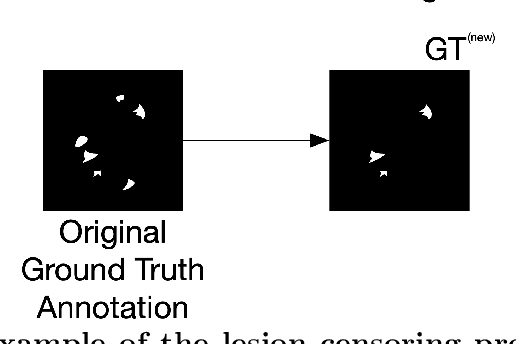
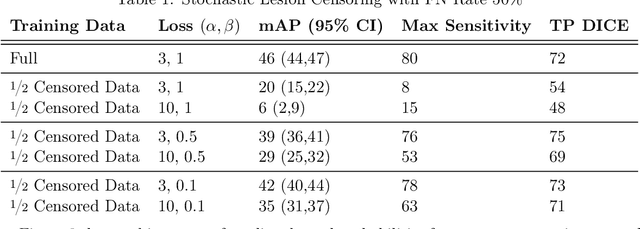
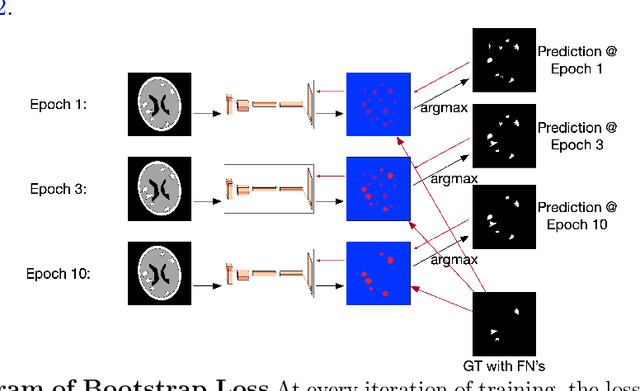
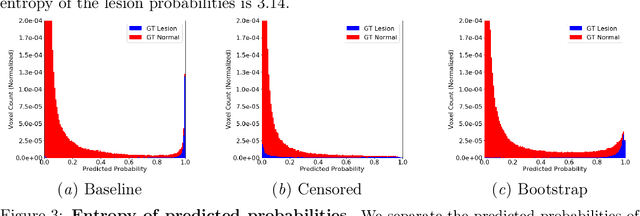
Abstract:Deep learning has proven to be an essential tool for medical image analysis. However, the need for accurately labeled input data, often requiring time- and labor-intensive annotation by experts, is a major limitation to the use of deep learning. One solution to this challenge is to allow for use of coarse or noisy labels, which could permit more efficient and scalable labeling of images. In this work, we develop a lopsided loss function based on entropy regularization that assumes the existence of a nontrivial false negative rate in the target annotations. Starting with a carefully annotated brain metastasis lesion dataset, we simulate data with false negatives by (1) randomly censoring the annotated lesions and (2) systematically censoring the smallest lesions. The latter better models true physician error because smaller lesions are harder to notice than the larger ones. Even with a simulated false negative rate as high as 50%, applying our loss function to randomly censored data preserves maximum sensitivity at 97% of the baseline with uncensored training data, compared to just 10% for a standard loss function. For the size-based censorship, performance is restored from 17% with the current standard to 88% with our lopsided bootstrap loss. Our work will enable more efficient scaling of the image labeling process, in parallel with other approaches on creating more efficient user interfaces and tools for annotation.
Handling Missing MRI Input Data in Deep Learning Segmentation of Brain Metastases: A Multi-Center Study
Dec 27, 2019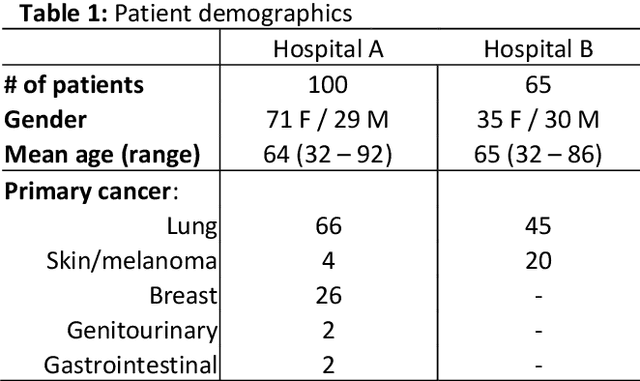
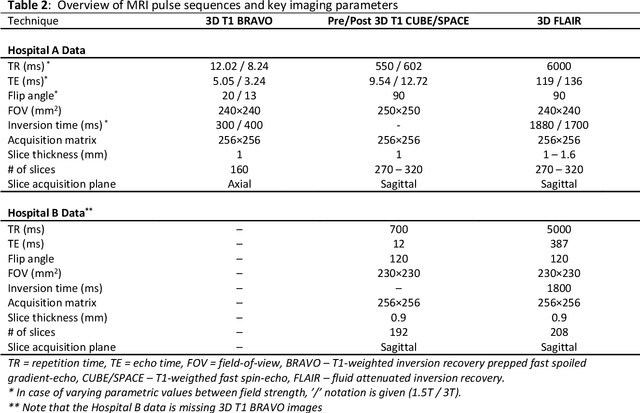
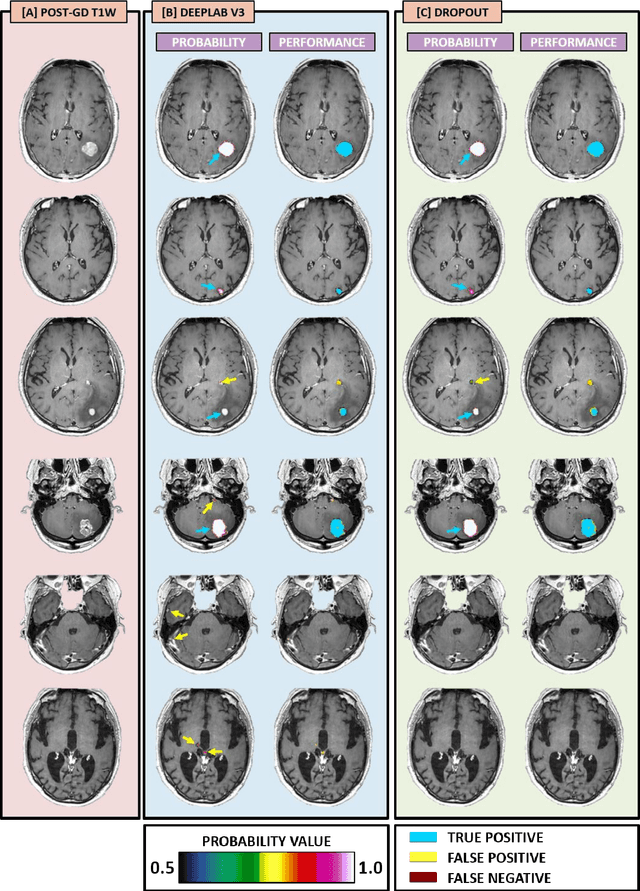
Abstract:The purpose was to assess the clinical value of a novel DropOut model for detecting and segmenting brain metastases, in which a neural network is trained on four distinct MRI sequences using an input dropout layer, thus simulating the scenario of missing MRI data by training on the full set and all possible subsets of the input data. This retrospective, multi-center study, evaluated 165 patients with brain metastases. A deep learning based segmentation model for automatic segmentation of brain metastases, named DropOut, was trained on multi-sequence MRI from 100 patients, and validated/tested on 10/55 patients. The segmentation results were compared with the performance of a state-of-the-art DeepLabV3 model. The MR sequences in the training set included pre- and post-gadolinium (Gd) T1-weighted 3D fast spin echo, post-Gd T1-weighted inversion recovery (IR) prepped fast spoiled gradient echo, and 3D fluid attenuated inversion recovery (FLAIR), whereas the test set did not include the IR prepped image-series. The ground truth were established by experienced neuroradiologists. The results were evaluated using precision, recall, Dice score, and receiver operating characteristics (ROC) curve statistics, while the Wilcoxon rank sum test was used to compare the performance of the two neural networks. The area under the ROC curve (AUC), averaged across all test cases, was 0.989+-0.029 for the DropOut model and 0.989+-0.023 for the DeepLabV3 model (p=0.62). The DropOut model showed a significantly higher Dice score compared to the DeepLabV3 model (0.795+-0.105 vs. 0.774+-0.104, p=0.017), and a significantly lower average false positive rate of 3.6/patient vs. 7.0/patient (p<0.001) using a 10mm3 lesion-size limit. The DropOut model may facilitate accurate detection and segmentation of brain metastases on a multi-center basis, even when the test cohort is missing MRI input data.
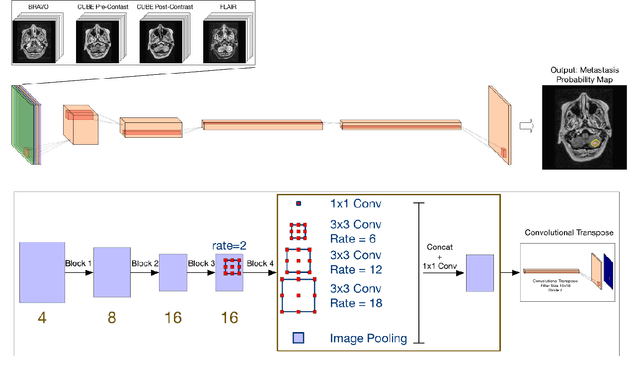
MRI Pulse Sequence Integration for Deep-Learning Based Brain Metastasis Segmentation
Dec 18, 2019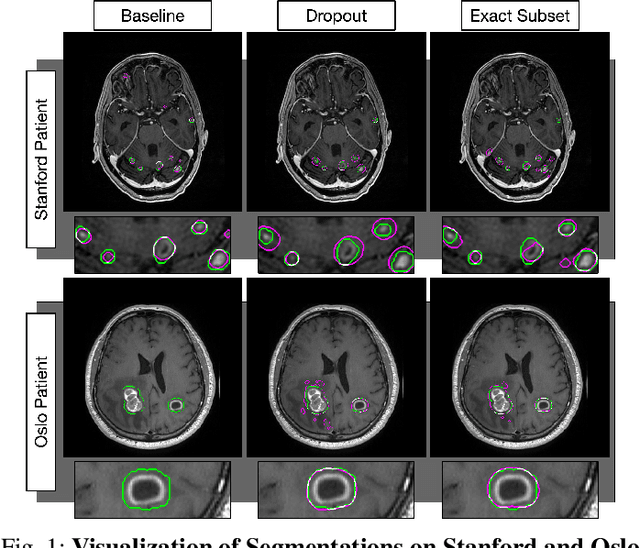
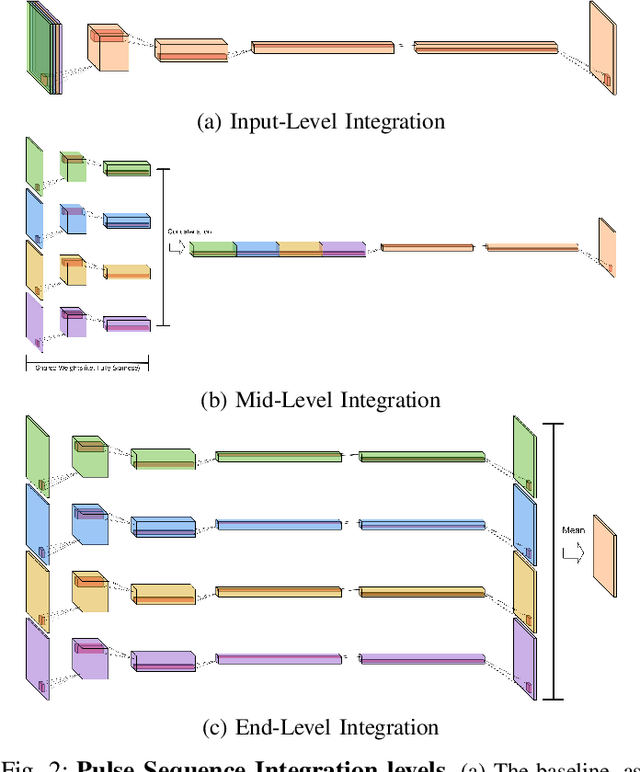
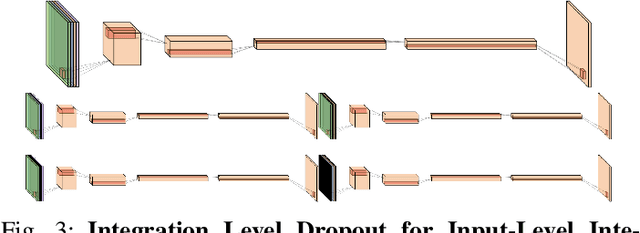
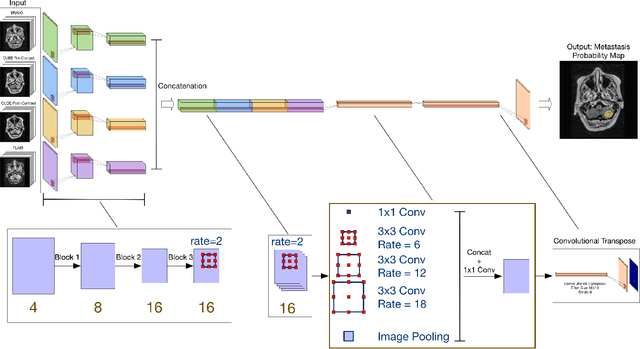
Abstract:Magnetic resonance (MR) imaging is an essential diagnostic tool in clinical medicine. Recently, a variety of deep learning methods have been applied to segmentation tasks in medical images, with promising results for computer-aided diagnosis. For MR images, effectively integrating different pulse sequences is important to optimize performance. However, the best way to integrate different pulse sequences remains unclear. In this study, we evaluate multiple architectural features and characterize their effects in the task of metastasis segmentation. Specifically, we consider (1) different pulse sequence integration schemas, (2) different modes of weight sharing for parallel network branches, and (3) a new approach for enabling robustness to missing pulse sequences. We find that levels of integration and modes of weight sharing that favor low variance work best in our regime of small data (n = 100). By adding an input-level dropout layer, we could preserve the overall performance of these networks while allowing for inference on inputs with missing pulse sequence. We illustrate not only the generalizability of the network but also the utility of this robustness when applying the trained model to data from a different center, which does not use the same pulse sequences. Finally, we apply network visualization methods to better understand which input features are most important for network performance. Together, these results provide a framework for building networks with enhanced robustness to missing data while maintaining comparable performance in medical imaging applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge