Chaoyu Chen
CoBa: Convergence Balancer for Multitask Finetuning of Large Language Models
Oct 09, 2024Abstract:Multi-task learning (MTL) benefits the fine-tuning of large language models (LLMs) by providing a single model with improved performance and generalization ability across tasks, presenting a resource-efficient alternative to developing separate models for each task. Yet, existing MTL strategies for LLMs often fall short by either being computationally intensive or failing to ensure simultaneous task convergence. This paper presents CoBa, a new MTL approach designed to effectively manage task convergence balance with minimal computational overhead. Utilizing Relative Convergence Scores (RCS), Absolute Convergence Scores (ACS), and a Divergence Factor (DF), CoBa dynamically adjusts task weights during the training process, ensuring that the validation loss of all tasks progress towards convergence at an even pace while mitigating the issue of individual task divergence. The results of our experiments involving three disparate datasets underscore that this approach not only fosters equilibrium in task improvement but enhances the LLMs' performance by up to 13% relative to the second-best baselines. Code is open-sourced at https://github.com/codefuse-ai/MFTCoder.
SQLfuse: Enhancing Text-to-SQL Performance through Comprehensive LLM Synergy
Jul 19, 2024



Abstract:Text-to-SQL conversion is a critical innovation, simplifying the transition from complex SQL to intuitive natural language queries, especially significant given SQL's prevalence in the job market across various roles. The rise of Large Language Models (LLMs) like GPT-3.5 and GPT-4 has greatly advanced this field, offering improved natural language understanding and the ability to generate nuanced SQL statements. However, the potential of open-source LLMs in Text-to-SQL applications remains underexplored, with many frameworks failing to leverage their full capabilities, particularly in handling complex database queries and incorporating feedback for iterative refinement. Addressing these limitations, this paper introduces SQLfuse, a robust system integrating open-source LLMs with a suite of tools to enhance Text-to-SQL translation's accuracy and usability. SQLfuse features four modules: schema mining, schema linking, SQL generation, and a SQL critic module, to not only generate but also continuously enhance SQL query quality. Demonstrated by its leading performance on the Spider Leaderboard and deployment by Ant Group, SQLfuse showcases the practical merits of open-source LLMs in diverse business contexts.
A Survey on Language Models for Code
Nov 19, 2023



Abstract:In this work we systematically review the recent advancements in code processing with language models, covering 50+ models, 30+ evaluation tasks, 150+ datasets, and 550 related works. We break down code processing models into general language models represented by the GPT family and specialized models that are specifically pretrained on code, often with tailored objectives. We discuss the relations and differences between these models, and highlight the historical transition of code modeling from statistical models and RNNs to pretrained Transformers and LLMs, which is exactly the same course that had been taken by NLP. We also discuss code-specific features such as AST, CFG, and unit tests, along with their application in training code language models, and identify key challenges and potential future directions in this domain. We keep the survey open and updated on GitHub repository at https://github.com/codefuse-ai/Awesome-Code-LLM.
MFTCoder: Boosting Code LLMs with Multitask Fine-Tuning
Nov 04, 2023Abstract:Code LLMs have emerged as a specialized research field, with remarkable studies dedicated to enhancing model's coding capabilities through fine-tuning on pre-trained models. Previous fine-tuning approaches were typically tailored to specific downstream tasks or scenarios, which meant separate fine-tuning for each task, requiring extensive training resources and posing challenges in terms of deployment and maintenance. Furthermore, these approaches failed to leverage the inherent interconnectedness among different code-related tasks. To overcome these limitations, we present a multi-task fine-tuning framework, MFTcoder, that enables simultaneous and parallel fine-tuning on multiple tasks. By incorporating various loss functions, we effectively address common challenges in multi-task learning, such as data imbalance, varying difficulty levels, and inconsistent convergence speeds. Extensive experiments have conclusively demonstrated that our multi-task fine-tuning approach outperforms both individual fine-tuning on single tasks and fine-tuning on a mixed ensemble of tasks. Moreover, MFTcoder offers efficient training capabilities, including efficient data tokenization modes and PEFT fine-tuning, resulting in significantly improved speed compared to traditional fine-tuning methods. MFTcoder seamlessly integrates with several mainstream open-source LLMs, such as CodeLLama and Qwen. Leveraging the CodeLLama foundation, our MFTcoder fine-tuned model, \textsc{CodeFuse-CodeLLama-34B}, achieves an impressive pass@1 score of 74.4\% on the HumaneEval benchmark, surpassing GPT-4 performance (67\%, zero-shot). MFTCoder is open-sourced at \url{https://github.com/codefuse-ai/MFTCOder}
FetusMapV2: Enhanced Fetal Pose Estimation in 3D Ultrasound
Oct 30, 2023Abstract:Fetal pose estimation in 3D ultrasound (US) involves identifying a set of associated fetal anatomical landmarks. Its primary objective is to provide comprehensive information about the fetus through landmark connections, thus benefiting various critical applications, such as biometric measurements, plane localization, and fetal movement monitoring. However, accurately estimating the 3D fetal pose in US volume has several challenges, including poor image quality, limited GPU memory for tackling high dimensional data, symmetrical or ambiguous anatomical structures, and considerable variations in fetal poses. In this study, we propose a novel 3D fetal pose estimation framework (called FetusMapV2) to overcome the above challenges. Our contribution is three-fold. First, we propose a heuristic scheme that explores the complementary network structure-unconstrained and activation-unreserved GPU memory management approaches, which can enlarge the input image resolution for better results under limited GPU memory. Second, we design a novel Pair Loss to mitigate confusion caused by symmetrical and similar anatomical structures. It separates the hidden classification task from the landmark localization task and thus progressively eases model learning. Last, we propose a shape priors-based self-supervised learning by selecting the relatively stable landmarks to refine the pose online. Extensive experiments and diverse applications on a large-scale fetal US dataset including 1000 volumes with 22 landmarks per volume demonstrate that our method outperforms other strong competitors.
CodeFuse-13B: A Pretrained Multi-lingual Code Large Language Model
Oct 10, 2023Abstract:Code Large Language Models (Code LLMs) have gained significant attention in the industry due to their wide applications in the full lifecycle of software engineering. However, the effectiveness of existing models in understanding non-English inputs for multi-lingual code-related tasks is still far from well studied. This paper introduces CodeFuse-13B, an open-sourced pre-trained code LLM. It is specifically designed for code-related tasks with both English and Chinese prompts and supports over 40 programming languages. CodeFuse achieves its effectiveness by utilizing a high quality pre-training dataset that is carefully filtered by program analyzers and optimized during the training process. Extensive experiments are conducted using real-world usage scenarios, the industry-standard benchmark HumanEval-x, and the specially designed CodeFuseEval for Chinese prompts. To assess the effectiveness of CodeFuse, we actively collected valuable human feedback from the AntGroup's software development process where CodeFuse has been successfully deployed. The results demonstrate that CodeFuse-13B achieves a HumanEval pass@1 score of 37.10%, positioning it as one of the top multi-lingual code LLMs with similar parameter sizes. In practical scenarios, such as code generation, code translation, code comments, and testcase generation, CodeFuse performs better than other models when confronted with Chinese prompts.
FFPN: Fourier Feature Pyramid Network for Ultrasound Image Segmentation
Aug 26, 2023



Abstract:Ultrasound (US) image segmentation is an active research area that requires real-time and highly accurate analysis in many scenarios. The detect-to-segment (DTS) frameworks have been recently proposed to balance accuracy and efficiency. However, existing approaches may suffer from inadequate contour encoding or fail to effectively leverage the encoded results. In this paper, we introduce a novel Fourier-anchor-based DTS framework called Fourier Feature Pyramid Network (FFPN) to address the aforementioned issues. The contributions of this paper are two fold. First, the FFPN utilizes Fourier Descriptors to adequately encode contours. Specifically, it maps Fourier series with similar amplitudes and frequencies into the same layer of the feature map, thereby effectively utilizing the encoded Fourier information. Second, we propose a Contour Sampling Refinement (CSR) module based on the contour proposals and refined features produced by the FFPN. This module extracts rich features around the predicted contours to further capture detailed information and refine the contours. Extensive experimental results on three large and challenging datasets demonstrate that our method outperforms other DTS methods in terms of accuracy and efficiency. Furthermore, our framework can generalize well to other detection or segmentation tasks.
Segment Anything Model for Medical Images?
May 01, 2023



Abstract:The Segment Anything Model (SAM) is the first foundation model for general image segmentation. It designed a novel promotable segmentation task, ensuring zero-shot image segmentation using the pre-trained model via two main modes including automatic everything and manual prompt. SAM has achieved impressive results on various natural image segmentation tasks. However, medical image segmentation (MIS) is more challenging due to the complex modalities, fine anatomical structures, uncertain and complex object boundaries, and wide-range object scales. Meanwhile, zero-shot and efficient MIS can well reduce the annotation time and boost the development of medical image analysis. Hence, SAM seems to be a potential tool and its performance on large medical datasets should be further validated. We collected and sorted 52 open-source datasets, and built a large medical segmentation dataset with 16 modalities, 68 objects, and 553K slices. We conducted a comprehensive analysis of different SAM testing strategies on the so-called COSMOS 553K dataset. Extensive experiments validate that SAM performs better with manual hints like points and boxes for object perception in medical images, leading to better performance in prompt mode compared to everything mode. Additionally, SAM shows remarkable performance in some specific objects and modalities, but is imperfect or even totally fails in other situations. Finally, we analyze the influence of different factors (e.g., the Fourier-based boundary complexity and size of the segmented objects) on SAM's segmentation performance. Extensive experiments validate that SAM's zero-shot segmentation capability is not sufficient to ensure its direct application to the MIS.
BALANCE: Bayesian Linear Attribution for Root Cause Localization
Jan 31, 2023



Abstract:Root Cause Analysis (RCA) plays an indispensable role in distributed data system maintenance and operations, as it bridges the gap between fault detection and system recovery. Existing works mainly study multidimensional localization or graph-based root cause localization. This paper opens up the possibilities of exploiting the recently developed framework of explainable AI (XAI) for the purpose of RCA. In particular, we propose BALANCE (BAyesian Linear AttributioN for root CausE localization), which formulates the problem of RCA through the lens of attribution in XAI and seeks to explain the anomalies in the target KPIs by the behavior of the candidate root causes. BALANCE consists of three innovative components. First, we propose a Bayesian multicollinear feature selection (BMFS) model to predict the target KPIs given the candidate root causes in a forward manner while promoting sparsity and concurrently paying attention to the correlation between the candidate root causes. Second, we introduce attribution analysis to compute the attribution score for each candidate in a backward manner. Third, we merge the estimated root causes related to each KPI if there are multiple KPIs. We extensively evaluate the proposed BALANCE method on one synthesis dataset as well as three real-world RCA tasks, that is, bad SQL localization, container fault localization, and fault type diagnosis for Exathlon. Results show that BALANCE outperforms the state-of-the-art (SOTA) methods in terms of accuracy with the least amount of running time, and achieves at least $6\%$ notably higher accuracy than SOTA methods for real tasks. BALANCE has been deployed to production to tackle real-world RCA problems, and the online results further advocate its usage for real-time diagnosis in distributed data systems.
Fine-grained Correlation Loss for Regression
Jul 01, 2022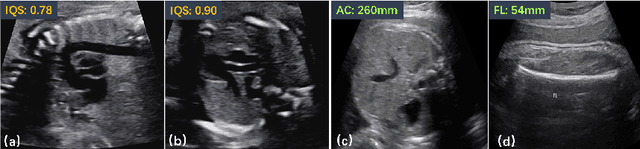
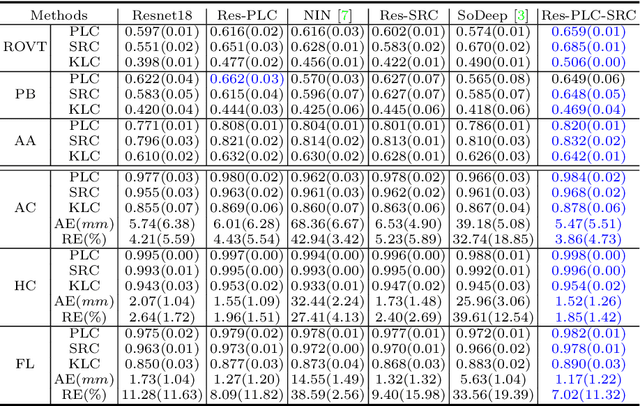
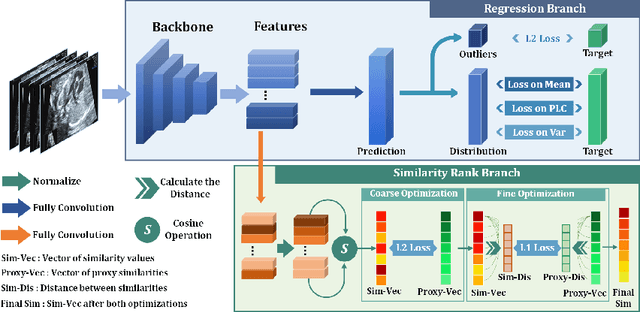
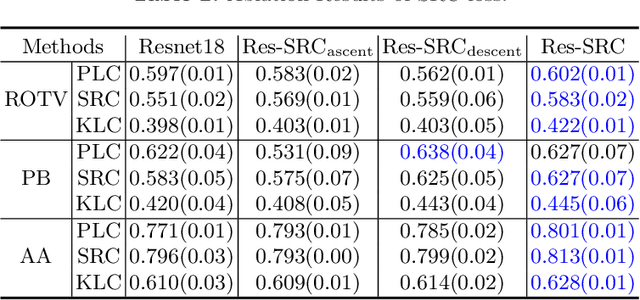
Abstract:Regression learning is classic and fundamental for medical image analysis. It provides the continuous mapping for many critical applications, like the attribute estimation, object detection, segmentation and non-rigid registration. However, previous studies mainly took the case-wise criteria, like the mean square errors, as the optimization objectives. They ignored the very important population-wise correlation criterion, which is exactly the final evaluation metric in many tasks. In this work, we propose to revisit the classic regression tasks with novel investigations on directly optimizing the fine-grained correlation losses. We mainly explore two complementary correlation indexes as learnable losses: Pearson linear correlation (PLC) and Spearman rank correlation (SRC). The contributions of this paper are two folds. First, for the PLC on global level, we propose a strategy to make it robust against the outliers and regularize the key distribution factors. These efforts significantly stabilize the learning and magnify the efficacy of PLC. Second, for the SRC on local level, we propose a coarse-to-fine scheme to ease the learning of the exact ranking order among samples. Specifically, we convert the learning for the ranking of samples into the learning of similarity relationships among samples. We extensively validate our method on two typical ultrasound image regression tasks, including the image quality assessment and bio-metric measurement. Experiments prove that, with the fine-grained guidance in directly optimizing the correlation, the regression performances are significantly improved. Our proposed correlation losses are general and can be extended to more important applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge