Carola-Bibiane Schönlieb
on behalf of the AIX-COVNET collaboration
Simultaneous Semantic and Instance Segmentation for Colon Nuclei Identification and Counting
Mar 01, 2022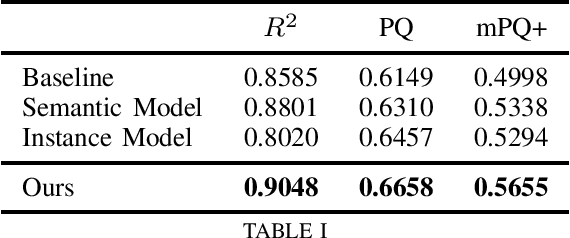
Abstract:We address the problem of automated nuclear segmentation, classification, and quantification from Haematoxylin and Eosin stained histology images, which is of great relevance for several downstream computational pathology applications. In this work, we present a solution framed as a simultaneous semantic and instance segmentation framework. Our solution is part of the Colon Nuclei Identification and Counting (CoNIC) Challenge. We first train a semantic and instance segmentation model separately. Our framework uses as backbone HoverNet and Cascade Mask-RCNN models. We then ensemble the results with a custom Non-Maximum Suppression embedding (NMS). In our framework, the semantic model computes a class prediction for the cells whilst the instance model provides a refined segmentation. We demonstrate, through our experimental results, that our model outperforms the provided baselines by a large margin.
Collaborative learning of images and geometrics for predicting isocitrate dehydrogenase status of glioma
Jan 14, 2022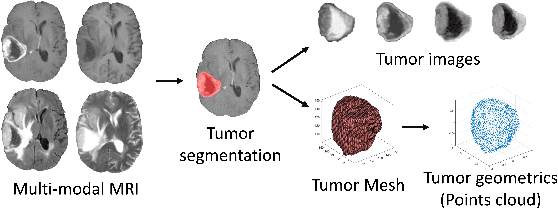
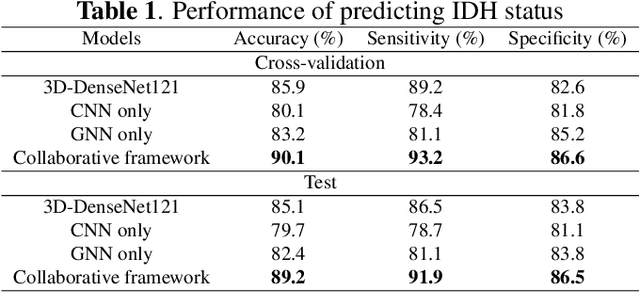
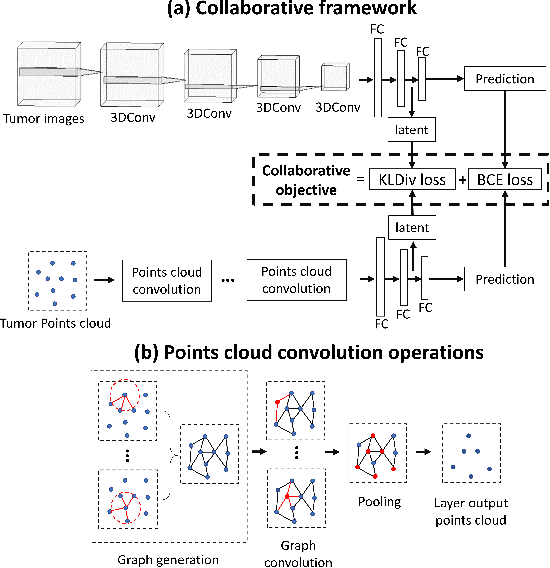
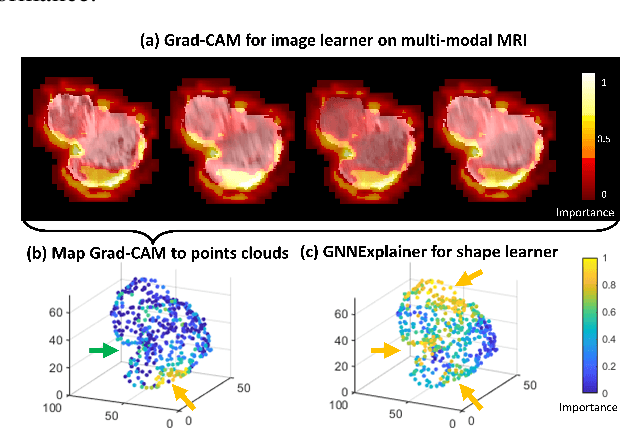
Abstract:The isocitrate dehydrogenase (IDH) gene mutation status is an important biomarker for glioma patients. The gold standard of IDH mutation detection requires tumour tissue obtained via invasive approaches and is usually expensive. Recent advancement in radiogenomics provides a non-invasive approach for predicting IDH mutation based on MRI. Meanwhile, tumor geometrics encompass crucial information for tumour phenotyping. Here we propose a collaborative learning framework that learns both tumor images and tumor geometrics using convolutional neural networks (CNN) and graph neural networks (GNN), respectively. Our results show that the proposed model outperforms the baseline model of 3D-DenseNet121. Further, the collaborative learning model achieves better performance than either the CNN or the GNN alone. The model interpretation shows that the CNN and GNN could identify common and unique regions of interest for IDH mutation prediction. In conclusion, collaborating image and geometric learners provides a novel approach for predicting genotype and characterising glioma.
AI-based Reconstruction for Fast MRI -- A Systematic Review and Meta-analysis
Dec 23, 2021
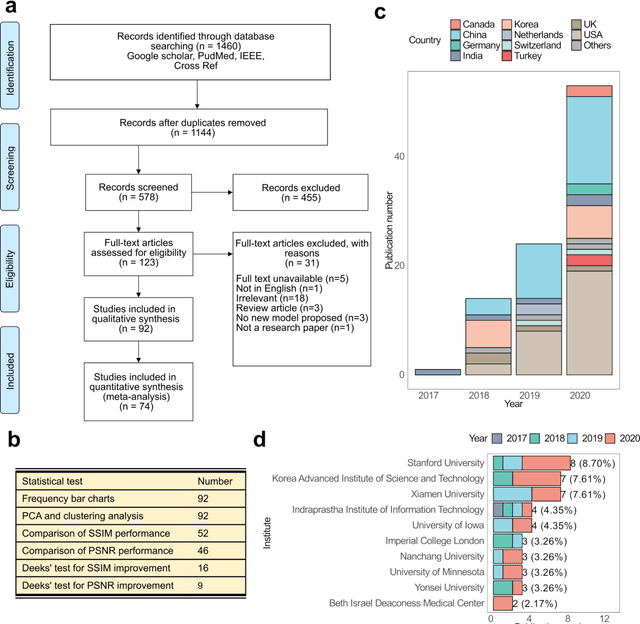
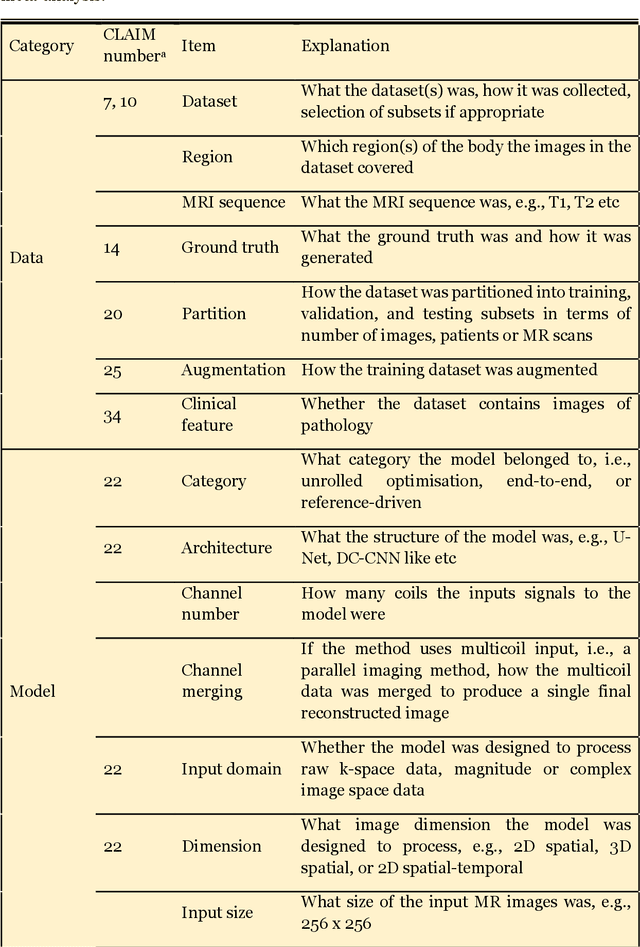
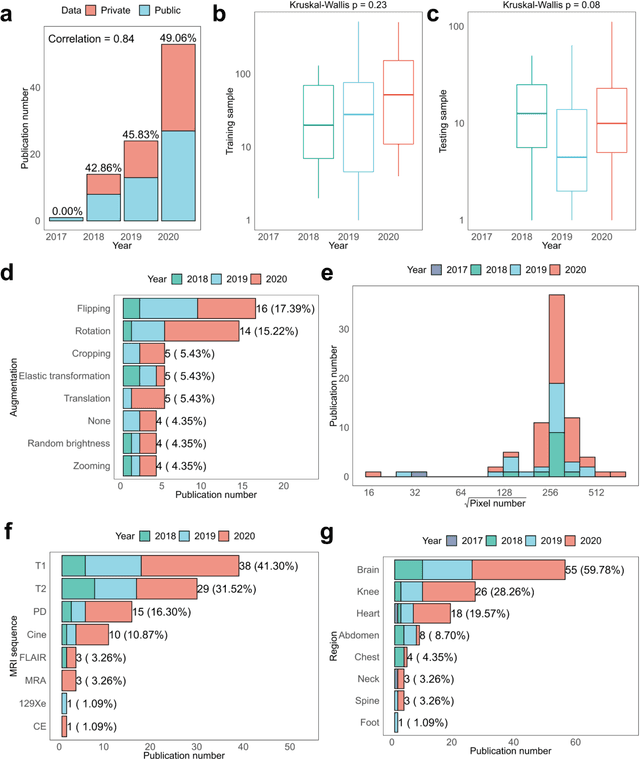
Abstract:Compressed sensing (CS) has been playing a key role in accelerating the magnetic resonance imaging (MRI) acquisition process. With the resurgence of artificial intelligence, deep neural networks and CS algorithms are being integrated to redefine the state of the art of fast MRI. The past several years have witnessed substantial growth in the complexity, diversity, and performance of deep learning-based CS techniques that are dedicated to fast MRI. In this meta-analysis, we systematically review the deep learning-based CS techniques for fast MRI, describe key model designs, highlight breakthroughs, and discuss promising directions. We have also introduced a comprehensive analysis framework and a classification system to assess the pivotal role of deep learning in CS-based acceleration for MRI.
A Continuous-time Stochastic Gradient Descent Method for Continuous Data
Dec 07, 2021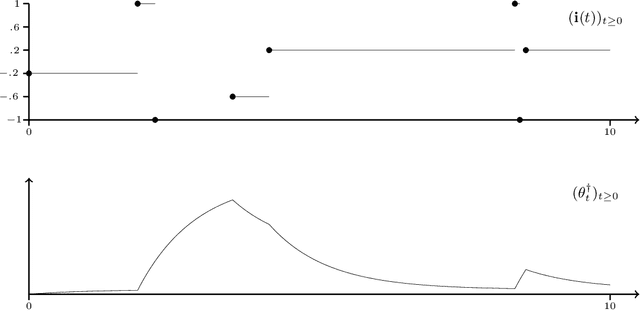
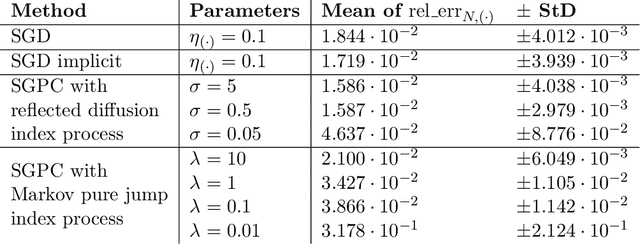
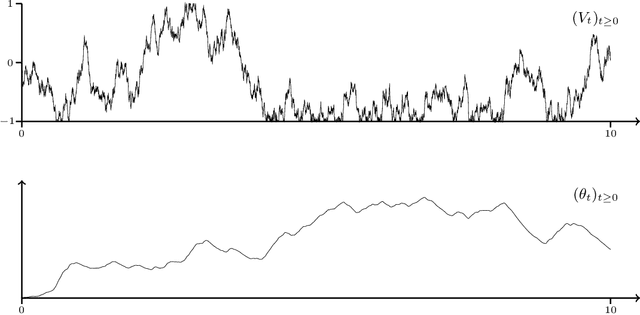
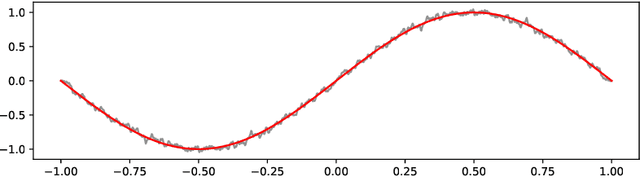
Abstract:Optimization problems with continuous data appear in, e.g., robust machine learning, functional data analysis, and variational inference. Here, the target function is given as an integral over a family of (continuously) indexed target functions - integrated with respect to a probability measure. Such problems can often be solved by stochastic optimization methods: performing optimization steps with respect to the indexed target function with randomly switched indices. In this work, we study a continuous-time variant of the stochastic gradient descent algorithm for optimization problems with continuous data. This so-called stochastic gradient process consists in a gradient flow minimizing an indexed target function that is coupled with a continuous-time index process determining the index. Index processes are, e.g., reflected diffusions, pure jump processes, or other L\'evy processes on compact spaces. Thus, we study multiple sampling patterns for the continuous data space and allow for data simulated or streamed at runtime of the algorithm. We analyze the approximation properties of the stochastic gradient process and study its longtime behavior and ergodicity under constant and decreasing learning rates. We end with illustrating the applicability of the stochastic gradient process in a polynomial regression problem with noisy functional data, as well as in a physics-informed neural network.
Conditional Image Generation with Score-Based Diffusion Models
Nov 26, 2021
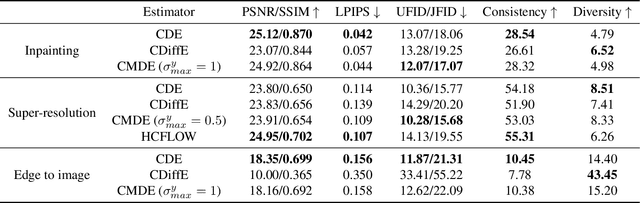
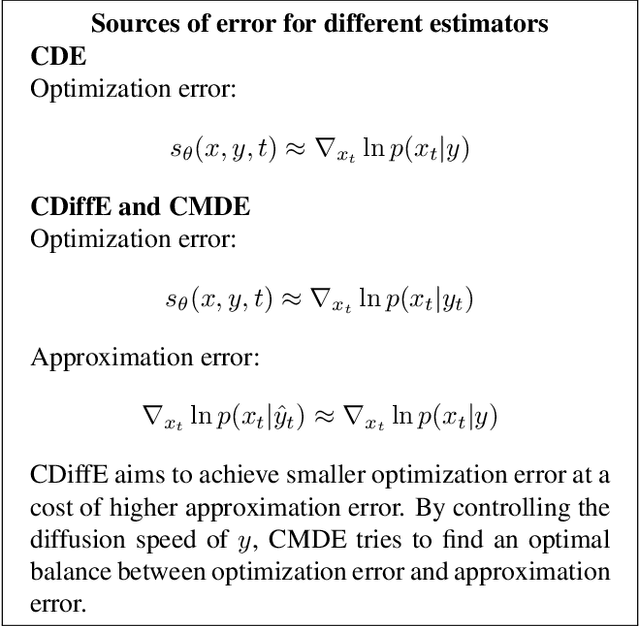
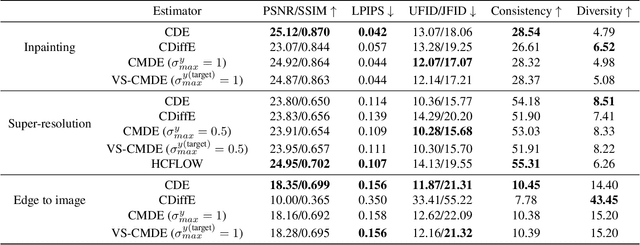
Abstract:Score-based diffusion models have emerged as one of the most promising frameworks for deep generative modelling. In this work we conduct a systematic comparison and theoretical analysis of different approaches to learning conditional probability distributions with score-based diffusion models. In particular, we prove results which provide a theoretical justification for one of the most successful estimators of the conditional score. Moreover, we introduce a multi-speed diffusion framework, which leads to a new estimator for the conditional score, performing on par with previous state-of-the-art approaches. Our theoretical and experimental findings are accompanied by an open source library MSDiff which allows for application and further research of multi-speed diffusion models.
Advancing COVID-19 Diagnosis with Privacy-Preserving Collaboration in Artificial Intelligence
Nov 18, 2021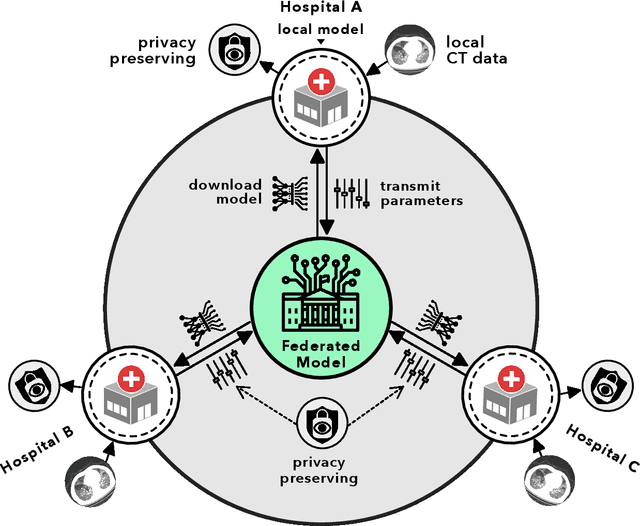
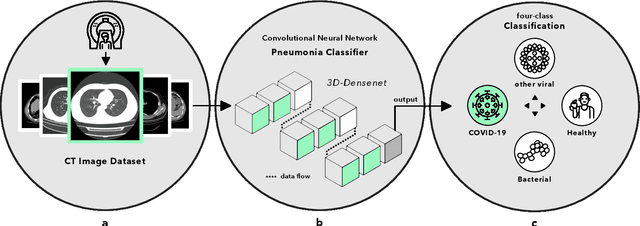
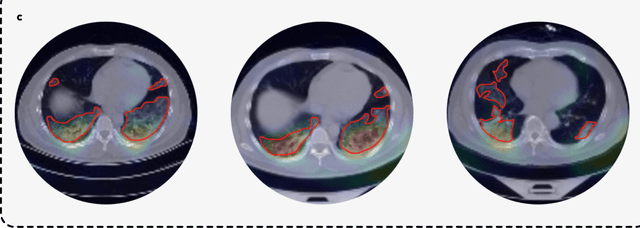
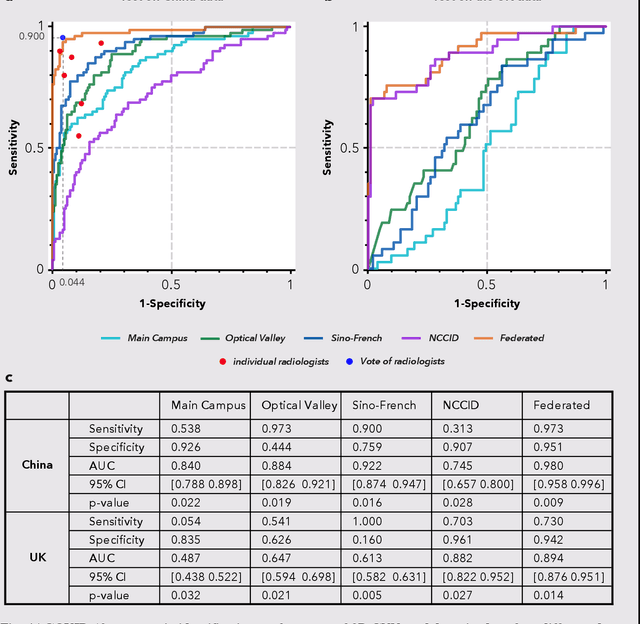
Abstract:Artificial intelligence (AI) provides a promising substitution for streamlining COVID-19 diagnoses. However, concerns surrounding security and trustworthiness impede the collection of large-scale representative medical data, posing a considerable challenge for training a well-generalised model in clinical practices. To address this, we launch the Unified CT-COVID AI Diagnostic Initiative (UCADI), where the AI model can be distributedly trained and independently executed at each host institution under a federated learning framework (FL) without data sharing. Here we show that our FL model outperformed all the local models by a large yield (test sensitivity /specificity in China: 0.973/0.951, in the UK: 0.730/0.942), achieving comparable performance with a panel of professional radiologists. We further evaluated the model on the hold-out (collected from another two hospitals leaving out the FL) and heterogeneous (acquired with contrast materials) data, provided visual explanations for decisions made by the model, and analysed the trade-offs between the model performance and the communication costs in the federated training process. Our study is based on 9,573 chest computed tomography scans (CTs) from 3,336 patients collected from 23 hospitals located in China and the UK. Collectively, our work advanced the prospects of utilising federated learning for privacy-preserving AI in digital health.
Focal Attention Networks: optimising attention for biomedical image segmentation
Oct 31, 2021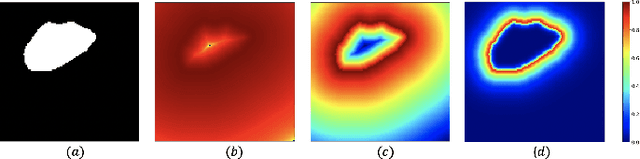
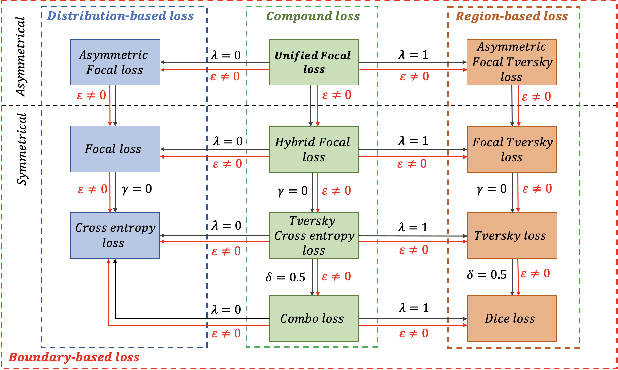
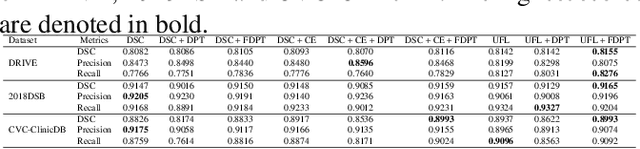
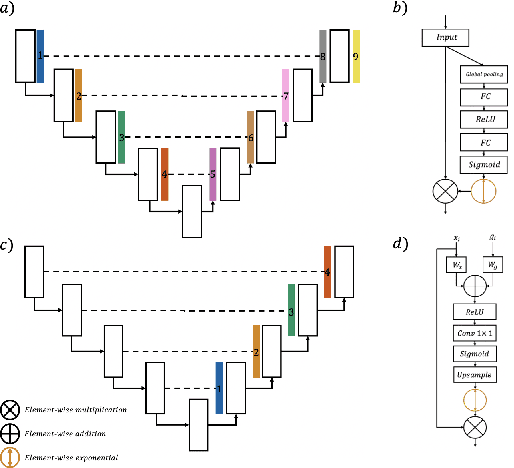
Abstract:In recent years, there has been increasing interest to incorporate attention into deep learning architectures for biomedical image segmentation. The modular design of attention mechanisms enables flexible integration into convolutional neural network architectures, such as the U-Net. Whether attention is appropriate to use, what type of attention to use, and where in the network to incorporate attention modules, are all important considerations that are currently overlooked. In this paper, we investigate the role of the Focal parameter in modulating attention, revealing a link between attention in loss functions and networks. By incorporating a Focal distance penalty term, we extend the Unified Focal loss framework to include boundary-based losses. Furthermore, we develop a simple and interpretable, dataset and model-specific heuristic to integrate the Focal parameter into the Squeeze-and-Excitation block and Attention Gate, achieving optimal performance with fewer number of attention modules on three well-validated biomedical imaging datasets, suggesting judicious use of attention modules results in better performance and efficiency.
Incorporating Boundary Uncertainty into loss functions for biomedical image segmentation
Oct 31, 2021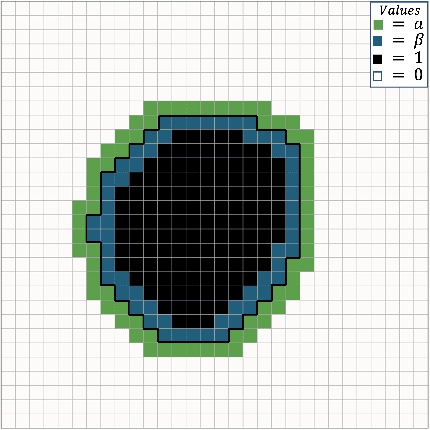

Abstract:Manual segmentation is used as the gold-standard for evaluating neural networks on automated image segmentation tasks. Due to considerable heterogeneity in shapes, colours and textures, demarcating object boundaries is particularly difficult in biomedical images, resulting in significant inter and intra-rater variability. Approaches, such as soft labelling and distance penalty term, apply a global transformation to the ground truth, redefining the loss function with respect to uncertainty. However, global operations are computationally expensive, and neither approach accurately reflects the uncertainty underlying manual annotation. In this paper, we propose the Boundary Uncertainty, which uses morphological operations to restrict soft labelling to object boundaries, providing an appropriate representation of uncertainty in ground truth labels, and may be adapted to enable robust model training where systematic manual segmentation errors are present. We incorporate Boundary Uncertainty with the Dice loss, achieving consistently improved performance across three well-validated biomedical imaging datasets compared to soft labelling and distance-weighted penalty. Boundary Uncertainty not only more accurately reflects the segmentation process, but it is also efficient, robust to segmentation errors and exhibits better generalisation.
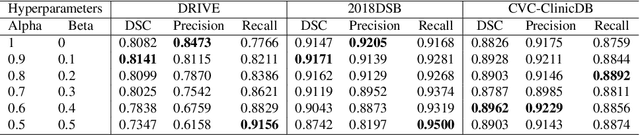
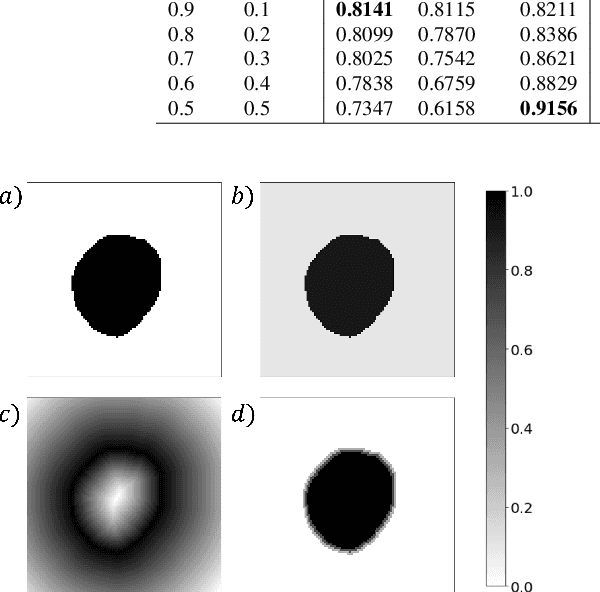
Calibrating the Dice loss to handle neural network overconfidence for biomedical image segmentation
Oct 31, 2021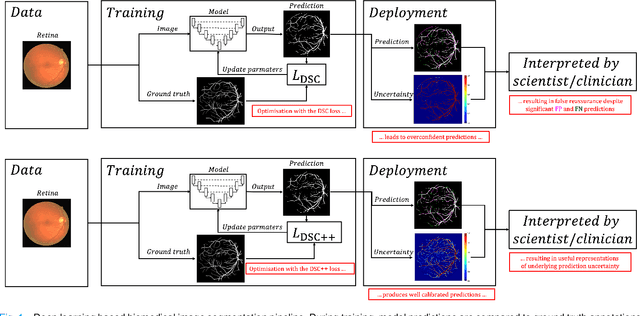
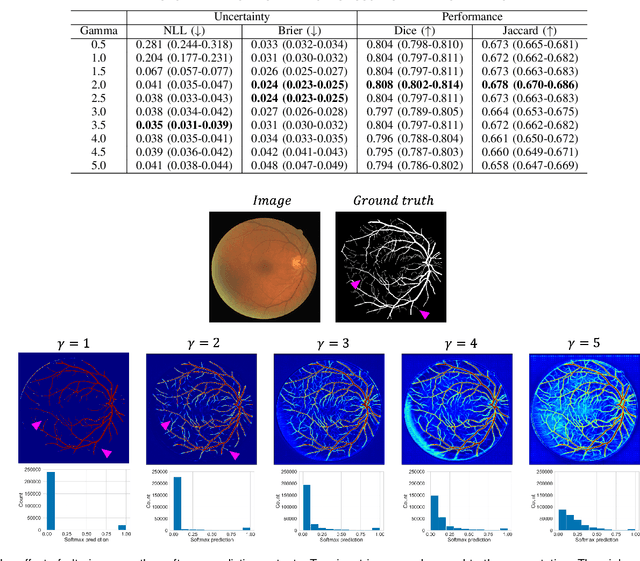
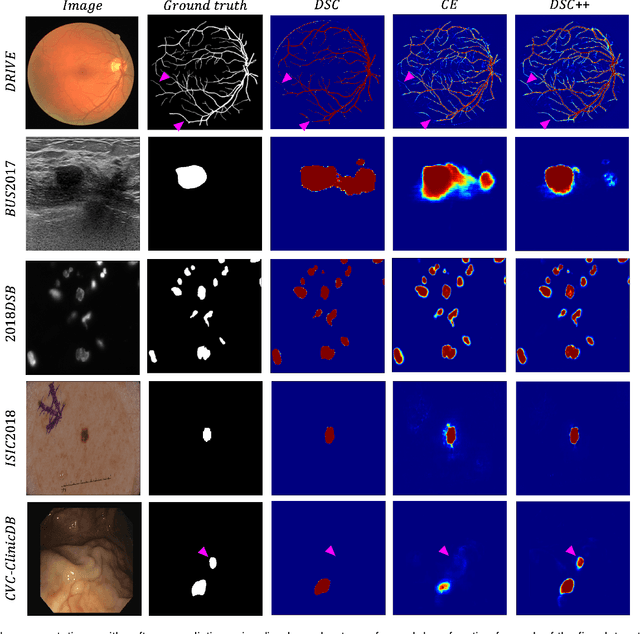
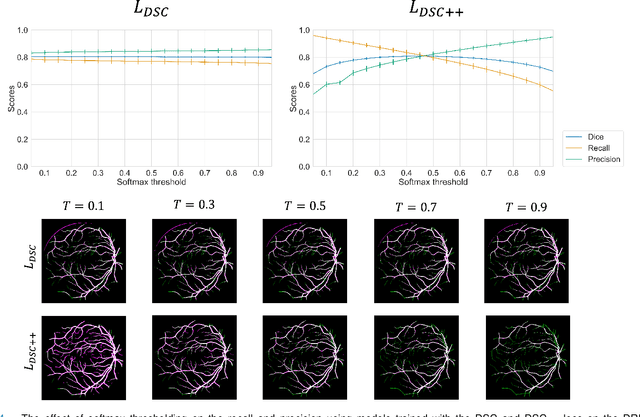
Abstract:The Dice similarity coefficient (DSC) is both a widely used metric and loss function for biomedical image segmentation due to its robustness to class imbalance. However, it is well known that the DSC loss is poorly calibrated, resulting in overconfident predictions that cannot be usefully interpreted in biomedical and clinical practice. Performance is often the only metric used to evaluate segmentations produced by deep neural networks, and calibration is often neglected. However, calibration is important for translation into biomedical and clinical practice, providing crucial contextual information to model predictions for interpretation by scientists and clinicians. In this study, we identify poor calibration as an emerging challenge of deep learning based biomedical image segmentation. We provide a simple yet effective extension of the DSC loss, named the DSC++ loss, that selectively modulates the penalty associated with overconfident, incorrect predictions. As a standalone loss function, the DSC++ loss achieves significantly improved calibration over the conventional DSC loss across five well-validated open-source biomedical imaging datasets. Similarly, we observe significantly improved when integrating the DSC++ loss into four DSC-based loss functions. Finally, we use softmax thresholding to illustrate that well calibrated outputs enable tailoring of precision-recall bias, an important post-processing technique to adapt the model predictions to suit the biomedical or clinical task. The DSC++ loss overcomes the major limitation of the DSC, providing a suitable loss function for training deep learning segmentation models for use in biomedical and clinical practice.
Learning convex regularizers satisfying the variational source condition for inverse problems
Oct 24, 2021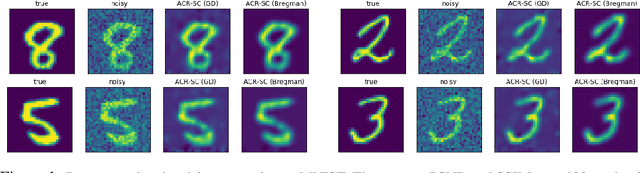
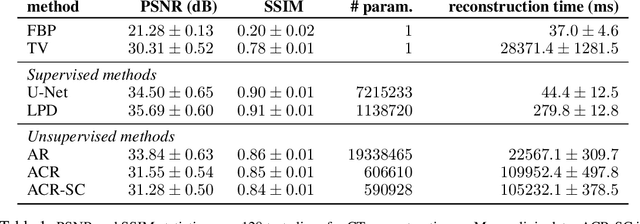
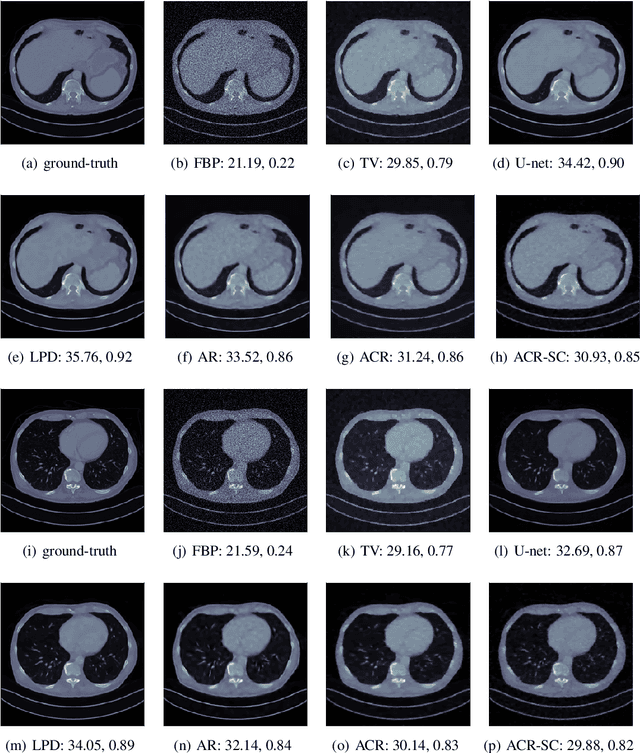
Abstract:Variational regularization has remained one of the most successful approaches for reconstruction in imaging inverse problems for several decades. With the emergence and astonishing success of deep learning in recent years, a considerable amount of research has gone into data-driven modeling of the regularizer in the variational setting. Our work extends a recently proposed method, referred to as adversarial convex regularization (ACR), that seeks to learn data-driven convex regularizers via adversarial training in an attempt to combine the power of data with the classical convex regularization theory. Specifically, we leverage the variational source condition (SC) during training to enforce that the ground-truth images minimize the variational loss corresponding to the learned convex regularizer. This is achieved by adding an appropriate penalty term to the ACR training objective. The resulting regularizer (abbreviated as ACR-SC) performs on par with the ACR, but unlike ACR, comes with a quantitative convergence rate estimate.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge