Alberto Gomez
Automatic retrieval of corresponding US views in longitudinal examinations
Jun 07, 2023



Abstract:Skeletal muscle atrophy is a common occurrence in critically ill patients in the intensive care unit (ICU) who spend long periods in bed. Muscle mass must be recovered through physiotherapy before patient discharge and ultrasound imaging is frequently used to assess the recovery process by measuring the muscle size over time. However, these manual measurements are subject to large variability, particularly since the scans are typically acquired on different days and potentially by different operators. In this paper, we propose a self-supervised contrastive learning approach to automatically retrieve similar ultrasound muscle views at different scan times. Three different models were compared using data from 67 patients acquired in the ICU. Results indicate that our contrastive model outperformed a supervised baseline model in the task of view retrieval with an AUC of 73.52% and when combined with an automatic segmentation model achieved 5.7%+/-0.24% error in cross-sectional area. Furthermore, a user study survey confirmed the efficacy of our model for muscle view retrieval.
Echo from noise: synthetic ultrasound image generation using diffusion models for real image segmentation
May 09, 2023Abstract:We propose a novel pipeline for the generation of synthetic images via Denoising Diffusion Probabilistic Models (DDPMs) guided by cardiac ultrasound semantic label maps. We show that these synthetic images can serve as a viable substitute for real data in the training of deep-learning models for medical image analysis tasks such as image segmentation. To demonstrate the effectiveness of this approach, we generated synthetic 2D echocardiography images and trained a neural network for segmentation of the left ventricle and left atrium. The performance of the network trained on exclusively synthetic images was evaluated on an unseen dataset of real images and yielded mean Dice scores of 88.5 $\pm 6.0$ , 92.3 $\pm 3.9$, 86.3 $\pm 10.7$ \% for left ventricular endocardial, epicardial and left atrial segmentation respectively. This represents an increase of $9.09$, $3.7$ and $15.0$ \% in Dice scores compared to the previous state-of-the-art. The proposed pipeline has the potential for application to a wide range of other tasks across various medical imaging modalities.
Feature-Conditioned Cascaded Video Diffusion Models for Precise Echocardiogram Synthesis
Mar 23, 2023Abstract:Image synthesis is expected to provide value for the translation of machine learning methods into clinical practice. Fundamental problems like model robustness, domain transfer, causal modelling, and operator training become approachable through synthetic data. Especially, heavily operator-dependant modalities like Ultrasound imaging require robust frameworks for image and video generation. So far, video generation has only been possible by providing input data that is as rich as the output data, e.g., image sequence plus conditioning in, video out. However, clinical documentation is usually scarce and only single images are reported and stored, thus retrospective patient-specific analysis or the generation of rich training data becomes impossible with current approaches. In this paper, we extend elucidated diffusion models for video modelling to generate plausible video sequences from single images and arbitrary conditioning with clinical parameters. We explore this idea within the context of echocardiograms by looking into the variation of the Left Ventricle Ejection Fraction, the most essential clinical metric gained from these examinations. We use the publicly available EchoNet-Dynamic dataset for all our experiments. Our image to sequence approach achieves an $R^2$ score of 93%, which is 38 points higher than recently proposed sequence to sequence generation methods. Code and models will be available at: https://github.com/HReynaud/EchoDiffusion.
Efficient Pix2Vox++ for 3D Cardiac Reconstruction from 2D echo views
Jul 27, 2022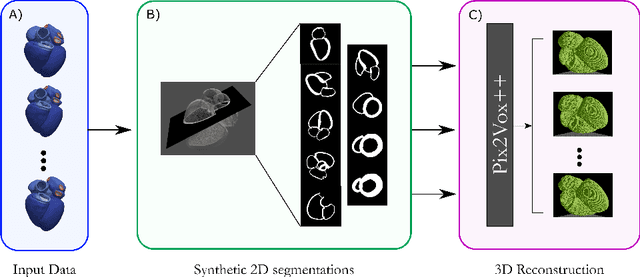
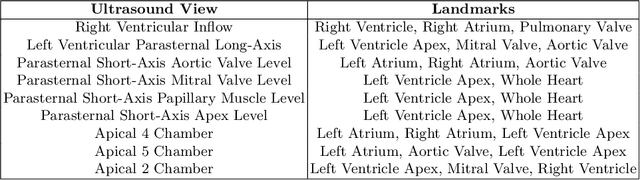
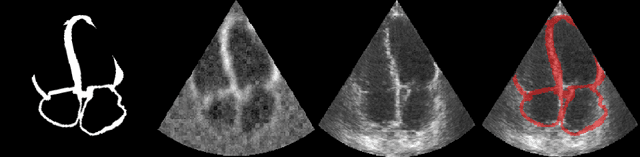
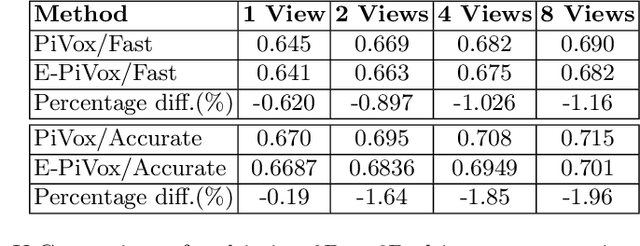
Abstract:Accurate geometric quantification of the human heart is a key step in the diagnosis of numerous cardiac diseases, and in the management of cardiac patients. Ultrasound imaging is the primary modality for cardiac imaging, however acquisition requires high operator skill, and its interpretation and analysis is difficult due to artifacts. Reconstructing cardiac anatomy in 3D can enable discovery of new biomarkers and make imaging less dependent on operator expertise, however most ultrasound systems only have 2D imaging capabilities. We propose both a simple alteration to the Pix2Vox++ networks for a sizeable reduction in memory usage and computational complexity, and a pipeline to perform reconstruction of 3D anatomy from 2D standard cardiac views, effectively enabling 3D anatomical reconstruction from limited 2D data. We evaluate our pipeline using synthetically generated data achieving accurate 3D whole-heart reconstructions (peak intersection over union score > 0.88) from just two standard anatomical 2D views of the heart. We also show preliminary results using real echo images.
Left Ventricle Contouring of Apical Three-Chamber Views on 2D Echocardiography
Jul 13, 2022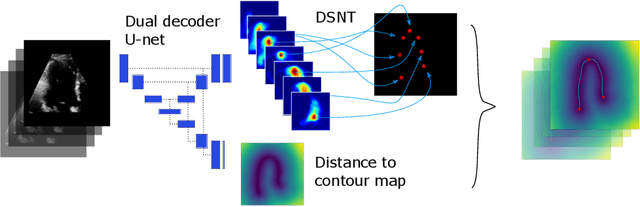
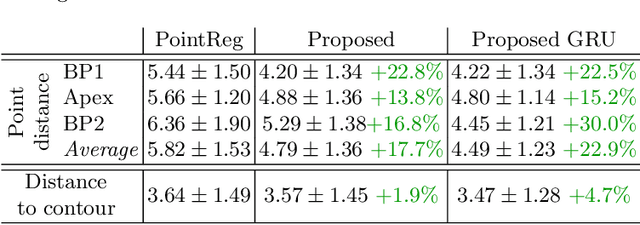
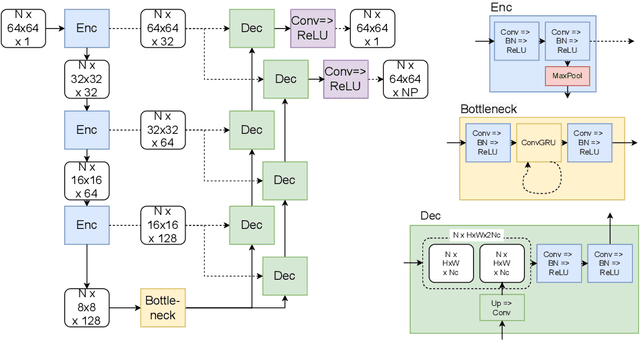
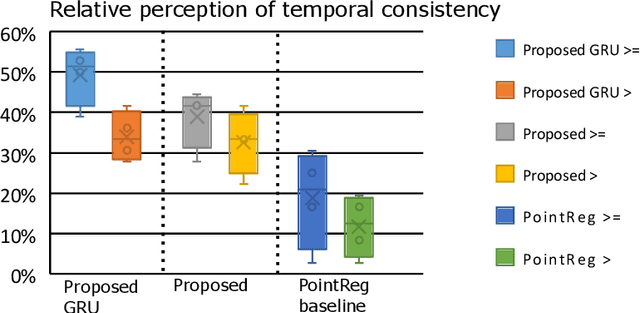
Abstract:We propose a new method to automatically contour the left ventricle on 2D echocardiographic images. Unlike most existing segmentation methods, which are based on predicting segmentation masks, we focus at predicting the endocardial contour and the key landmark points within this contour (basal points and apex). This provides a representation that is closer to how experts perform manual annotations and hence produce results that are physiologically more plausible. Our proposed method uses a two-headed network based on the U-Net architecture. One head predicts the 7 contour points, and the other head predicts a distance map to the contour. This approach was compared to the U-Net and to a point based approach, achieving performance gains of up to 30\% in terms of landmark localisation (<4.5mm) and distance to the ground truth contour (<3.5mm).
Placenta Segmentation in Ultrasound Imaging: Addressing Sources of Uncertainty and Limited Field-of-View
Jun 29, 2022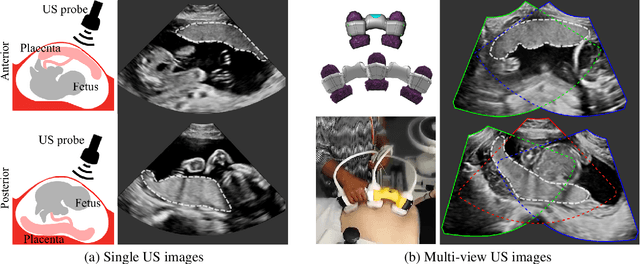
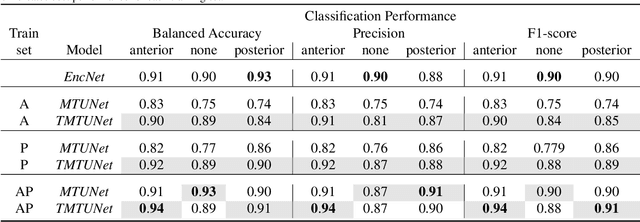
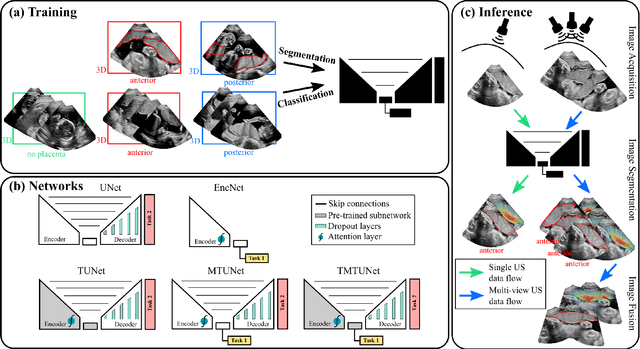
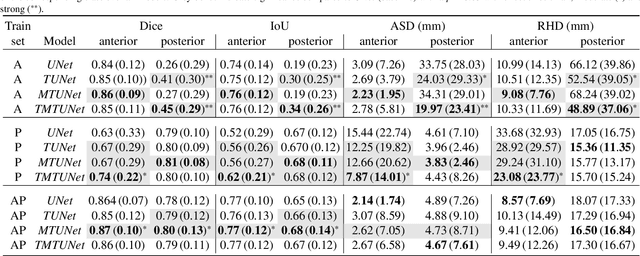
Abstract:Automatic segmentation of the placenta in fetal ultrasound (US) is challenging due to the (i) high diversity of placenta appearance, (ii) the restricted quality in US resulting in highly variable reference annotations, and (iii) the limited field-of-view of US prohibiting whole placenta assessment at late gestation. In this work, we address these three challenges with a multi-task learning approach that combines the classification of placental location (e.g., anterior, posterior) and semantic placenta segmentation in a single convolutional neural network. Through the classification task the model can learn from larger and more diverse datasets while improving the accuracy of the segmentation task in particular in limited training set conditions. With this approach we investigate the variability in annotations from multiple raters and show that our automatic segmentations (Dice of 0.86 for anterior and 0.83 for posterior placentas) achieve human-level performance as compared to intra- and inter-observer variability. Lastly, our approach can deliver whole placenta segmentation using a multi-view US acquisition pipeline consisting of three stages: multi-probe image acquisition, image fusion and image segmentation. This results in high quality segmentation of larger structures such as the placenta in US with reduced image artifacts which are beyond the field-of-view of single probes.
AI-enabled Assessment of Cardiac Systolic and Diastolic Function from Echocardiography
Mar 21, 2022
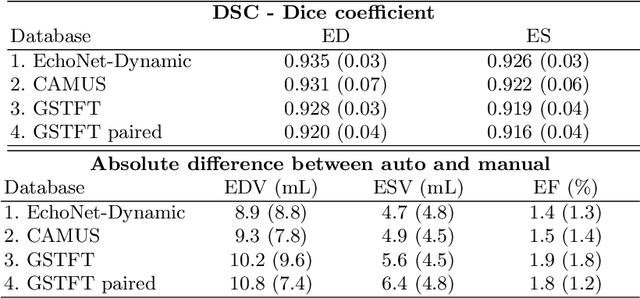
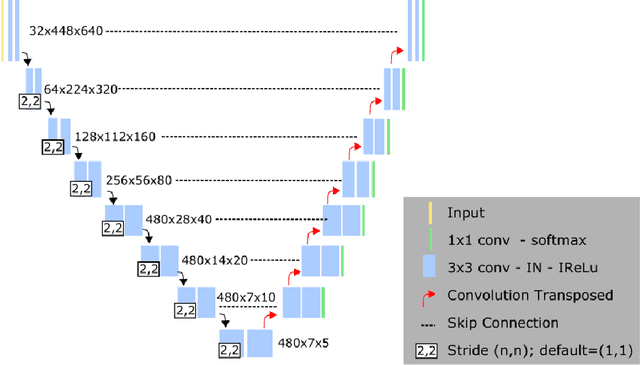
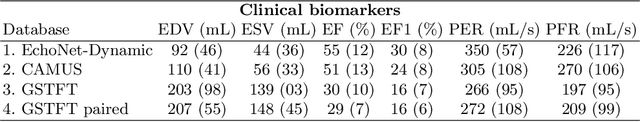
Abstract:Left ventricular (LV) function is an important factor in terms of patient management, outcome, and long-term survival of patients with heart disease. The most recently published clinical guidelines for heart failure recognise that over reliance on only one measure of cardiac function (LV ejection fraction) as a diagnostic and treatment stratification biomarker is suboptimal. Recent advances in AI-based echocardiography analysis have shown excellent results on automated estimation of LV volumes and LV ejection fraction. However, from time-varying 2-D echocardiography acquisition, a richer description of cardiac function can be obtained by estimating functional biomarkers from the complete cardiac cycle. In this work we propose for the first time an AI approach for deriving advanced biomarkers of systolic and diastolic LV function from 2-D echocardiography based on segmentations of the full cardiac cycle. These biomarkers will allow clinicians to obtain a much richer picture of the heart in health and disease. The AI model is based on the 'nn-Unet' framework and was trained and tested using four different databases. Results show excellent agreement between manual and automated analysis and showcase the potential of the advanced systolic and diastolic biomarkers for patient stratification. Finally, for a subset of 50 cases, we perform a correlation analysis between clinical biomarkers derived from echocardiography and CMR and we show excellent agreement between the two modalities.
B-line Detection in Lung Ultrasound Videos: Cartesian vs Polar Representation
Jul 26, 2021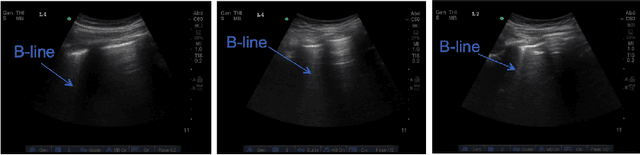

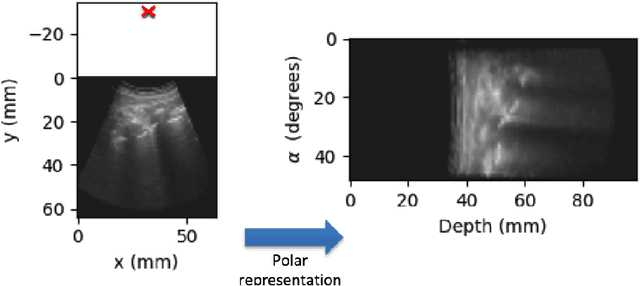
Abstract:Lung ultrasound (LUS) imaging is becoming popular in the intensive care units (ICU) for assessing lung abnormalities such as the appearance of B-line artefacts as a result of severe dengue. These artefacts appear in the LUS images and disappear quickly, making their manual detection very challenging. They also extend radially following the propagation of the sound waves. As a result, we hypothesize that a polar representation may be more adequate for automatic image analysis of these images. This paper presents an attention-based Convolutional+LSTM model to automatically detect B-lines in LUS videos, comparing performance when image data is taken in Cartesian and polar representations. Results indicate that the proposed framework with polar representation achieves competitive performance compared to the Cartesian representation for B-line classification and that attention mechanism can provide better localization.
Automatic Detection of B-lines in Lung Ultrasound Videos From Severe Dengue Patients
Feb 01, 2021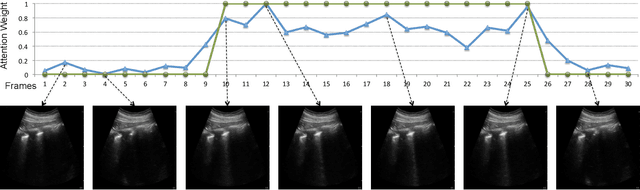
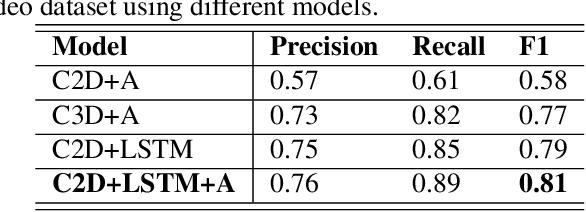
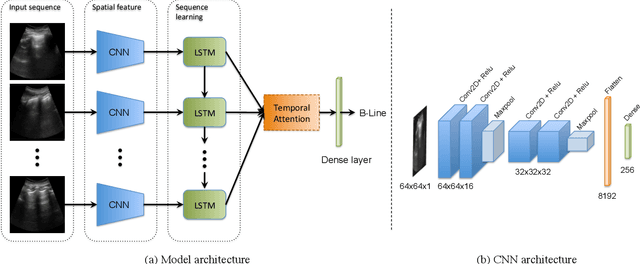
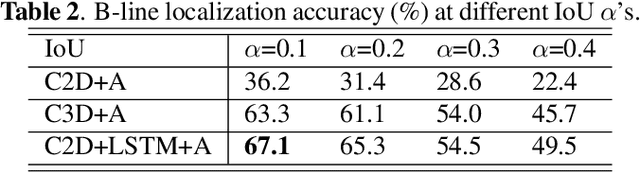
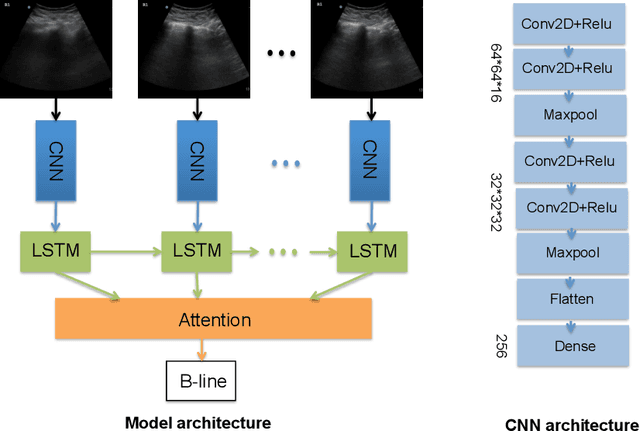
Abstract:Lung ultrasound (LUS) imaging is used to assess lung abnormalities, including the presence of B-line artefacts due to fluid leakage into the lungs caused by a variety of diseases. However, manual detection of these artefacts is challenging. In this paper, we propose a novel methodology to automatically detect and localize B-lines in LUS videos using deep neural networks trained with weak labels. To this end, we combine a convolutional neural network (CNN) with a long short-term memory (LSTM) network and a temporal attention mechanism. Four different models are compared using data from 60 patients. Results show that our best model can determine whether one-second clips contain B-lines or not with an F1 score of 0.81, and extracts a representative frame with B-lines with an accuracy of 87.5%.
Mutual Information-based Disentangled Neural Networks for Classifying Unseen Categories in Different Domains: Application to Fetal Ultrasound Imaging
Oct 30, 2020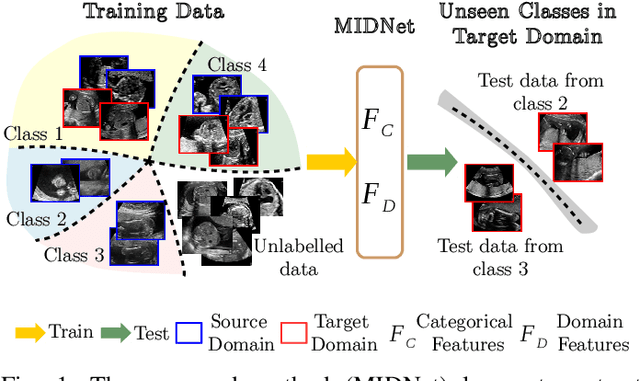
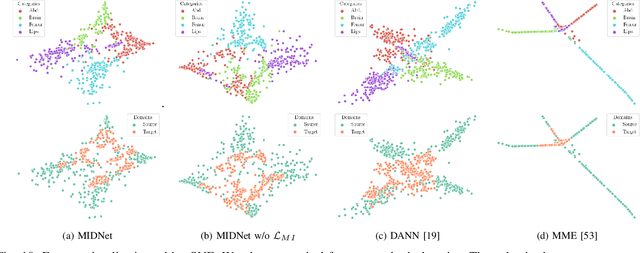
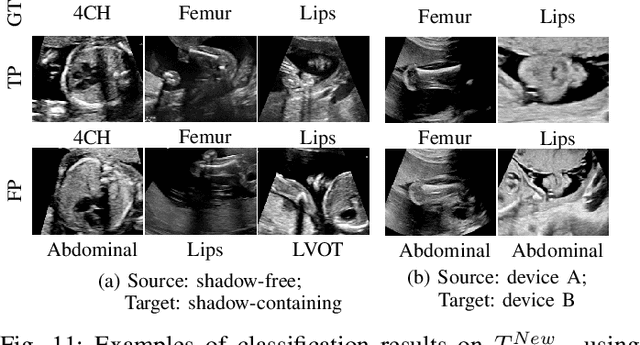
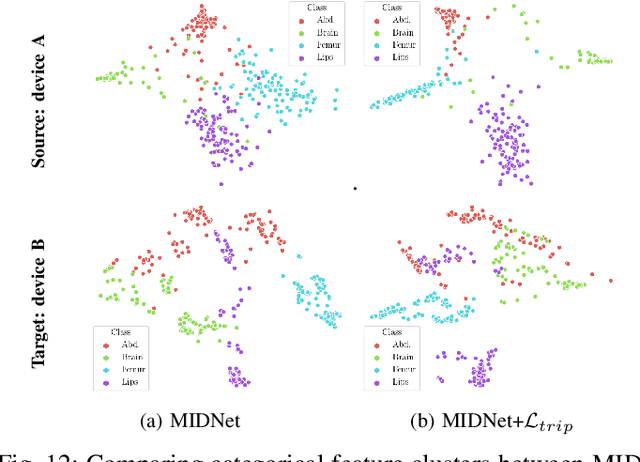
Abstract:Deep neural networks exhibit limited generalizability across images with different entangled domain features and categorical features. Learning generalizable features that can form universal categorical decision boundaries across domains is an interesting and difficult challenge. This problem occurs frequently in medical imaging applications when attempts are made to deploy and improve deep learning models across different image acquisition devices, across acquisition parameters or if some classes are unavailable in new training databases. To address this problem, we propose Mutual Information-based Disentangled Neural Networks (MIDNet), which extract generalizable categorical features to transfer knowledge to unseen categories in a target domain. The proposed MIDNet adopts a semi-supervised learning paradigm to alleviate the dependency on labeled data. This is important for real-world applications where data annotation is time-consuming, costly and requires training and expertise. We extensively evaluate the proposed method on fetal ultrasound datasets for two different image classification tasks where domain features are respectively defined by shadow artifacts and image acquisition devices. Experimental results show that the proposed method outperforms the state-of-the-art on the classification of unseen categories in a target domain with sparsely labeled training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge