Jacqueline Matthew
Whole-examination AI estimation of fetal biometrics from 20-week ultrasound scans
Jan 02, 2024Abstract:The current approach to fetal anomaly screening is based on biometric measurements derived from individually selected ultrasound images. In this paper, we introduce a paradigm shift that attains human-level performance in biometric measurement by aggregating automatically extracted biometrics from every frame across an entire scan, with no need for operator intervention. We use a convolutional neural network to classify each frame of an ultrasound video recording. We then measure fetal biometrics in every frame where appropriate anatomy is visible. We use a Bayesian method to estimate the true value of each biometric from a large number of measurements and probabilistically reject outliers. We performed a retrospective experiment on 1457 recordings (comprising 48 million frames) of 20-week ultrasound scans, estimated fetal biometrics in those scans and compared our estimates to the measurements sonographers took during the scan. Our method achieves human-level performance in estimating fetal biometrics and estimates well-calibrated credible intervals in which the true biometric value is expected to lie.
Towards Realistic Ultrasound Fetal Brain Imaging Synthesis
Apr 08, 2023
Abstract:Prenatal ultrasound imaging is the first-choice modality to assess fetal health. Medical image datasets for AI and ML methods must be diverse (i.e. diagnoses, diseases, pathologies, scanners, demographics, etc), however there are few public ultrasound fetal imaging datasets due to insufficient amounts of clinical data, patient privacy, rare occurrence of abnormalities in general practice, and limited experts for data collection and validation. To address such data scarcity, we proposed generative adversarial networks (GAN)-based models, diffusion-super-resolution-GAN and transformer-based-GAN, to synthesise images of fetal ultrasound brain planes from one public dataset. We reported that GAN-based methods can generate 256x256 pixel size of fetal ultrasound trans-cerebellum brain image plane with stable training losses, resulting in lower FID values for diffusion-super-resolution-GAN (average 7.04 and lower FID 5.09 at epoch 10) than the FID values of transformer-based-GAN (average 36.02 and lower 28.93 at epoch 60). The results of this work illustrate the potential of GAN-based methods to synthesise realistic high-resolution ultrasound images, leading to future work with other fetal brain planes, anatomies, devices and the need of a pool of experts to evaluate synthesised images. Code, data and other resources to reproduce this work are available at \url{https://github.com/budai4medtech/midl2023}.
Placenta Segmentation in Ultrasound Imaging: Addressing Sources of Uncertainty and Limited Field-of-View
Jun 29, 2022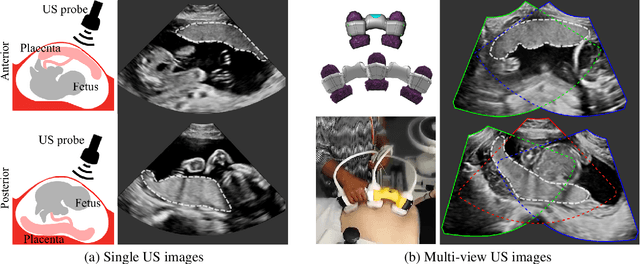
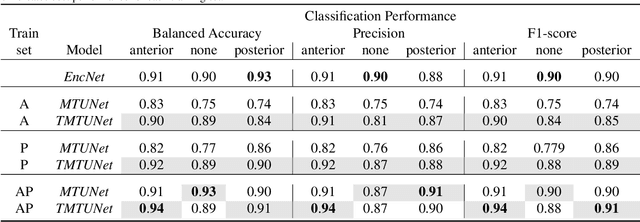
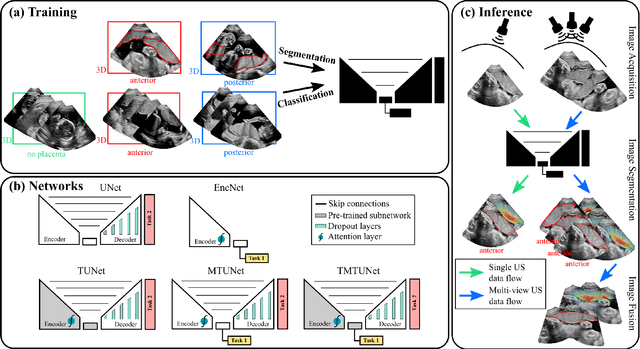
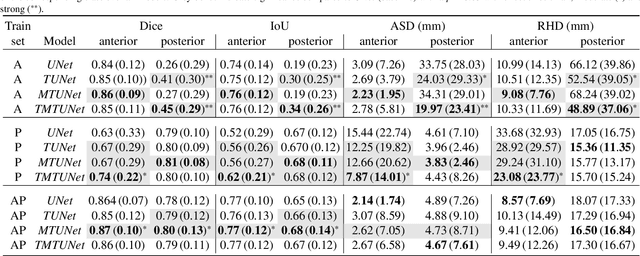
Abstract:Automatic segmentation of the placenta in fetal ultrasound (US) is challenging due to the (i) high diversity of placenta appearance, (ii) the restricted quality in US resulting in highly variable reference annotations, and (iii) the limited field-of-view of US prohibiting whole placenta assessment at late gestation. In this work, we address these three challenges with a multi-task learning approach that combines the classification of placental location (e.g., anterior, posterior) and semantic placenta segmentation in a single convolutional neural network. Through the classification task the model can learn from larger and more diverse datasets while improving the accuracy of the segmentation task in particular in limited training set conditions. With this approach we investigate the variability in annotations from multiple raters and show that our automatic segmentations (Dice of 0.86 for anterior and 0.83 for posterior placentas) achieve human-level performance as compared to intra- and inter-observer variability. Lastly, our approach can deliver whole placenta segmentation using a multi-view US acquisition pipeline consisting of three stages: multi-probe image acquisition, image fusion and image segmentation. This results in high quality segmentation of larger structures such as the placenta in US with reduced image artifacts which are beyond the field-of-view of single probes.
Empirical Study of Quality Image Assessment for Synthesis of Fetal Head Ultrasound Imaging with DCGANs
Jun 01, 2022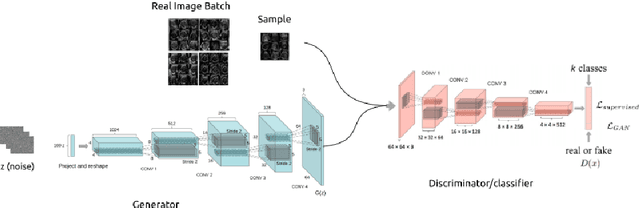
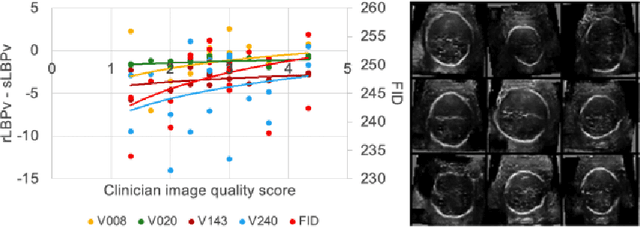
Abstract:In this work, we present an empirical study of DCGANs for synthetic generation of fetal head ultrasound, consisting of hyperparameter heuristics and image quality assessment. We present experiments to show the impact of different image sizes, epochs, data size input, and learning rates for quality image assessment on four metrics: mutual information (MI), fr\'echet inception distance (FID), peak-signal-to-noise ratio (PSNR), and local binary pattern vector (LBPv). The results show that FID and LBPv have stronger relationship with clinical image quality scores. The resources to reproduce this work are available at \url{https://github.com/xfetus/miua2022}.
Can non-specialists provide high quality gold standard labels in challenging modalities?
Jul 30, 2021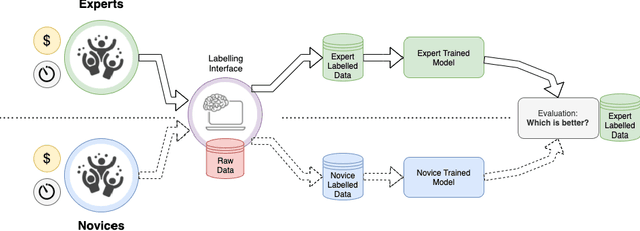
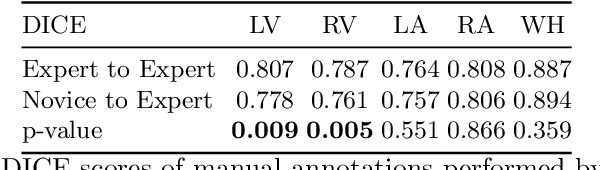
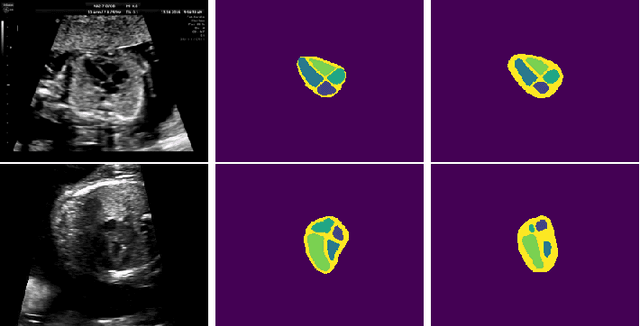
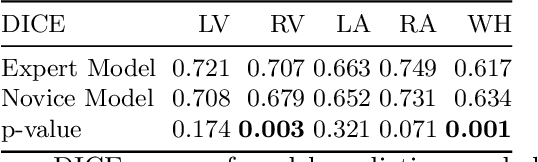
Abstract:Probably yes. -- Supervised Deep Learning dominates performance scores for many computer vision tasks and defines the state-of-the-art. However, medical image analysis lags behind natural image applications. One of the many reasons is the lack of well annotated medical image data available to researchers. One of the first things researchers are told is that we require significant expertise to reliably and accurately interpret and label such data. We see significant inter- and intra-observer variability between expert annotations of medical images. Still, it is a widely held assumption that novice annotators are unable to provide useful annotations for use by clinical Deep Learning models. In this work we challenge this assumption and examine the implications of using a minimally trained novice labelling workforce to acquire annotations for a complex medical image dataset. We study the time and cost implications of using novice annotators, the raw performance of novice annotators compared to gold-standard expert annotators, and the downstream effects on a trained Deep Learning segmentation model's performance for detecting a specific congenital heart disease (hypoplastic left heart syndrome) in fetal ultrasound imaging.
Learning normal appearance for fetal anomaly screening: Application to the unsupervised detection of Hypoplastic Left Heart Syndrome
Nov 15, 2020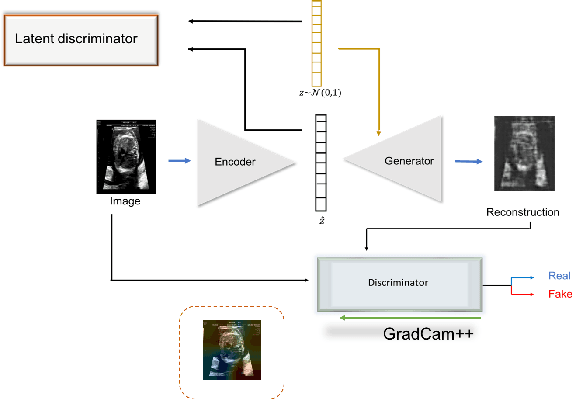
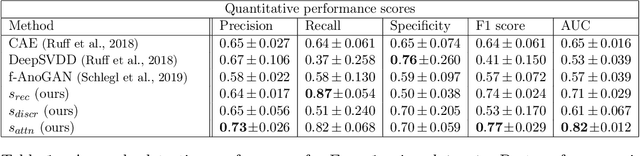
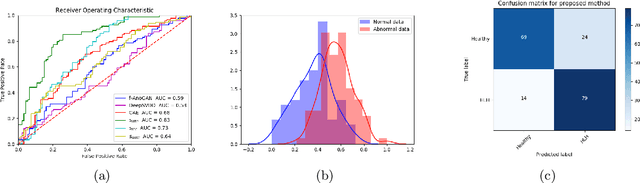
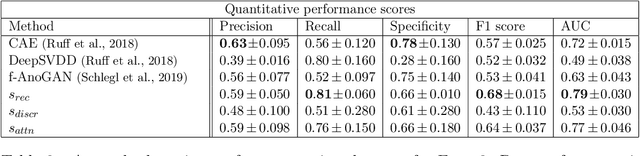
Abstract:Congenital heart disease is considered as one the most common groups of congenital malformations which affects $6-11$ per $1000$ newborns. In this work, an automated framework for detection of cardiac anomalies during ultrasound screening is proposed and evaluated on the example of Hypoplastic Left Heart Syndrome (HLHS), a sub-category of congenital heart disease. We propose an unsupervised approach that learns healthy anatomy exclusively from clinically confirmed normal control patients. We evaluate a number of known anomaly detection frameworks together with a new model architecture based on the $\alpha$-GAN network and find evidence that the proposed model performs significantly better than the state-of-the-art in image-based anomaly detection, yielding average $0.81$ AUC \emph{and} a better robustness towards initialisation compared to previous works.
Mutual Information-based Disentangled Neural Networks for Classifying Unseen Categories in Different Domains: Application to Fetal Ultrasound Imaging
Oct 30, 2020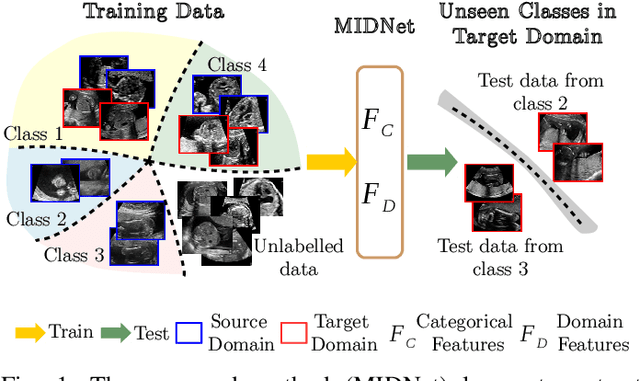
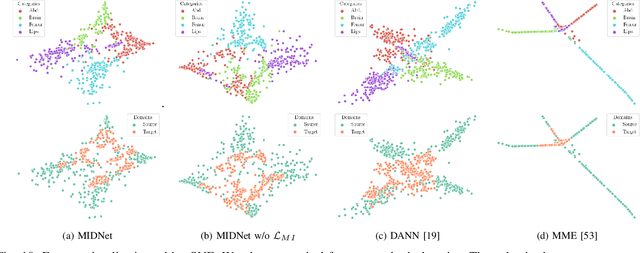
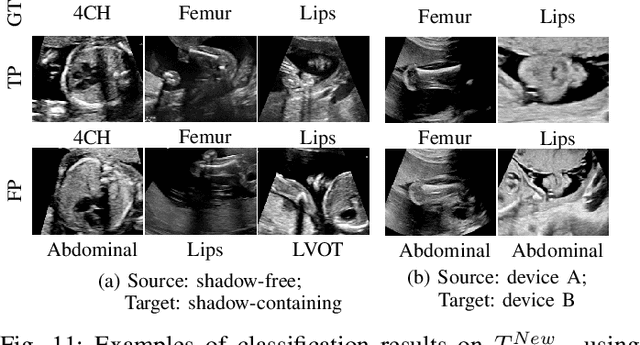
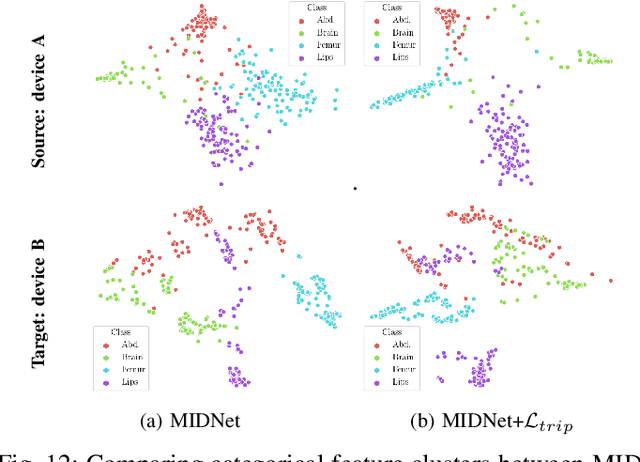
Abstract:Deep neural networks exhibit limited generalizability across images with different entangled domain features and categorical features. Learning generalizable features that can form universal categorical decision boundaries across domains is an interesting and difficult challenge. This problem occurs frequently in medical imaging applications when attempts are made to deploy and improve deep learning models across different image acquisition devices, across acquisition parameters or if some classes are unavailable in new training databases. To address this problem, we propose Mutual Information-based Disentangled Neural Networks (MIDNet), which extract generalizable categorical features to transfer knowledge to unseen categories in a target domain. The proposed MIDNet adopts a semi-supervised learning paradigm to alleviate the dependency on labeled data. This is important for real-world applications where data annotation is time-consuming, costly and requires training and expertise. We extensively evaluate the proposed method on fetal ultrasound datasets for two different image classification tasks where domain features are respectively defined by shadow artifacts and image acquisition devices. Experimental results show that the proposed method outperforms the state-of-the-art on the classification of unseen categories in a target domain with sparsely labeled training data.
Screen Tracking for Clinical Translation of Live Ultrasound Image Analysis Methods
Jul 13, 2020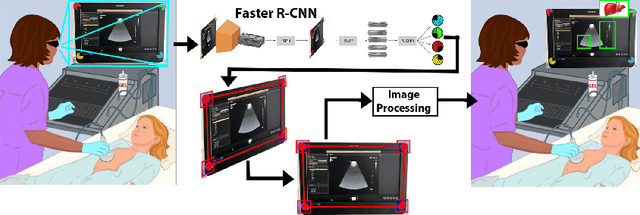
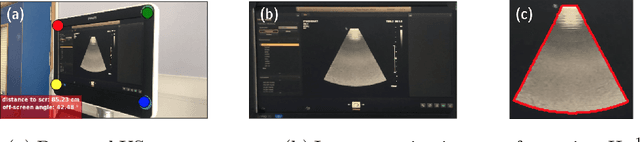
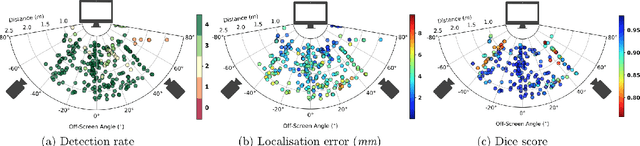
Abstract:Ultrasound (US) imaging is one of the most commonly used non-invasive imaging techniques. However, US image acquisition requires simultaneous guidance of the transducer and interpretation of images, which is a highly challenging task that requires years of training. Despite many recent developments in intra-examination US image analysis, the results are not easy to translate to a clinical setting. We propose a generic framework to extract the US images and superimpose the results of an analysis task, without any need for physical connection or alteration to the US system. The proposed method captures the US image by tracking the screen with a camera fixed at the sonographer's view point and reformats the captured image to the right aspect ratio, in 87.66 +- 3.73ms on average. It is hypothesized that this would enable to input such retrieved image into an image processing pipeline to extract information that can help improve the examination. This information could eventually be projected back to the sonographer's field of view in real time using, for example, an augmented reality (AR) headset.
Confident Head Circumference Measurement from Ultrasound with Real-time Feedback for Sonographers
Aug 07, 2019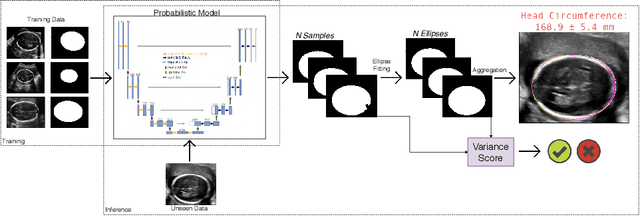
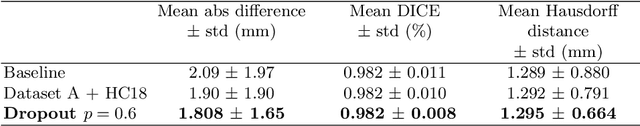
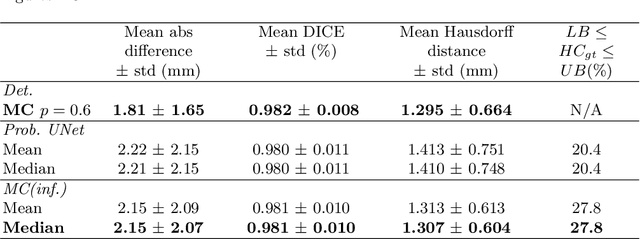
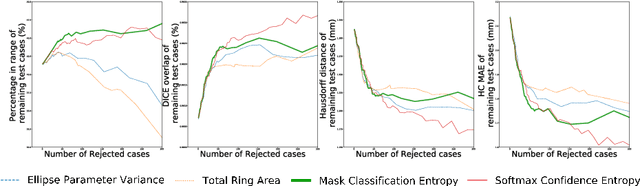
Abstract:Manual estimation of fetal Head Circumference (HC) from Ultrasound (US) is a key biometric for monitoring the healthy development of fetuses. Unfortunately, such measurements are subject to large inter-observer variability, resulting in low early-detection rates of fetal abnormalities. To address this issue, we propose a novel probabilistic Deep Learning approach for real-time automated estimation of fetal HC. This system feeds back statistics on measurement robustness to inform users how confident a deep neural network is in evaluating suitable views acquired during free-hand ultrasound examination. In real-time scenarios, this approach may be exploited to guide operators to scan planes that are as close as possible to the underlying distribution of training images, for the purpose of improving inter-operator consistency. We train on free-hand ultrasound data from over 2000 subjects (2848 training/540 test) and show that our method is able to predict HC measurements within 1.81$\pm$1.65mm deviation from the ground truth, with 50% of the test images fully contained within the predicted confidence margins, and an average of 1.82$\pm$1.78mm deviation from the margin for the remaining cases that are not fully contained.
Robotic-assisted Ultrasound for Fetal Imaging: Evolution from Single-arm to Dual-arm System
Apr 10, 2019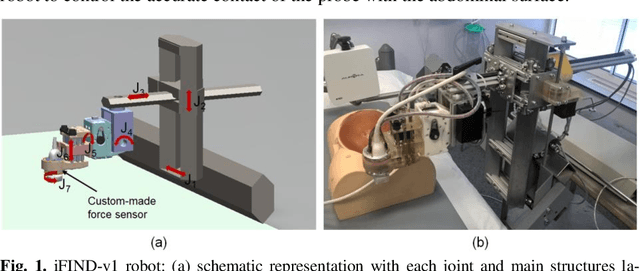
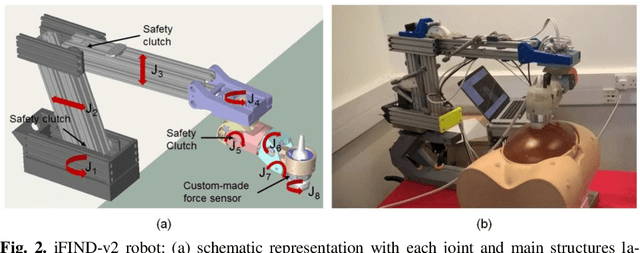
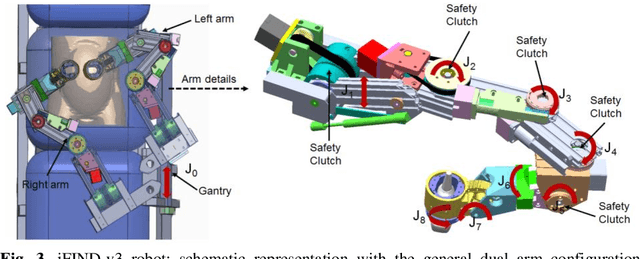
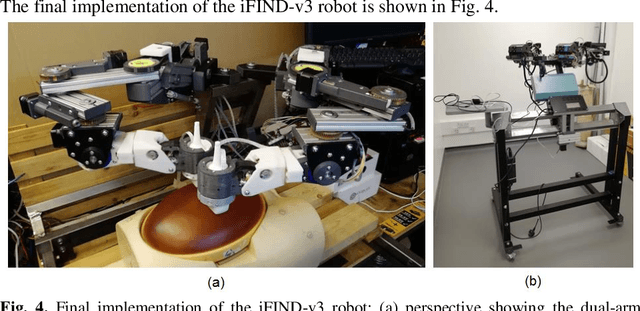
Abstract:The development of robotic-assisted extracorporeal ultrasound systems has a long history and a number of projects have been proposed since the 1990s focusing on different technical aspects. These aim to resolve the deficiencies of on-site manual manipulation of hand-held ultrasound probes. This paper presents the recent ongoing developments of a series of bespoke robotic systems, including both single-arm and dual-arm versions, for a project known as intelligent Fetal Imaging and Diagnosis (iFIND). After a brief review of the development history of the extracorporeal ultrasound robotic system used for fetal and abdominal examinations, the specific aim of the iFIND robots, the design evolution, the implementation details of each version, and the initial clinical feedback of the iFIND robot series are presented. Based on the preliminary testing of these newly-proposed robots on 42 volunteers, the successful and re-liable working of the mechatronic systems were validated. Analysis of a participant questionnaire indicates a comfortable scanning experience for the volunteers and a good acceptance rate to being scanned by the robots.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge