Uxio Hermida
Neural Implicit Heart Coordinates: 3D cardiac shape reconstruction from sparse segmentations
Dec 23, 2025Abstract:Accurate reconstruction of cardiac anatomy from sparse clinical images remains a major challenge in patient-specific modeling. While neural implicit functions have previously been applied to this task, their application to mapping anatomical consistency across subjects has been limited. In this work, we introduce Neural Implicit Heart Coordinates (NIHCs), a standardized implicit coordinate system, based on universal ventricular coordinates, that provides a common anatomical reference frame for the human heart. Our method predicts NIHCs directly from a limited number of 2D segmentations (sparse acquisition) and subsequently decodes them into dense 3D segmentations and high-resolution meshes at arbitrary output resolution. Trained on a large dataset of 5,000 cardiac meshes, the model achieves high reconstruction accuracy on clinical contours, with mean Euclidean surface errors of 2.51$\pm$0.33 mm in a diseased cohort (n=4549) and 2.3$\pm$0.36 mm in a healthy cohort (n=5576). The NIHC representation enables anatomically coherent reconstruction even under severe slice sparsity and segmentation noise, faithfully recovering complex structures such as the valve planes. Compared with traditional pipelines, inference time is reduced from over 60 s to 5-15 s. These results demonstrate that NIHCs constitute a robust and efficient anatomical representation for patient-specific 3D cardiac reconstruction from minimal input data.
Echo from noise: synthetic ultrasound image generation using diffusion models for real image segmentation
May 09, 2023Abstract:We propose a novel pipeline for the generation of synthetic images via Denoising Diffusion Probabilistic Models (DDPMs) guided by cardiac ultrasound semantic label maps. We show that these synthetic images can serve as a viable substitute for real data in the training of deep-learning models for medical image analysis tasks such as image segmentation. To demonstrate the effectiveness of this approach, we generated synthetic 2D echocardiography images and trained a neural network for segmentation of the left ventricle and left atrium. The performance of the network trained on exclusively synthetic images was evaluated on an unseen dataset of real images and yielded mean Dice scores of 88.5 $\pm 6.0$ , 92.3 $\pm 3.9$, 86.3 $\pm 10.7$ \% for left ventricular endocardial, epicardial and left atrial segmentation respectively. This represents an increase of $9.09$, $3.7$ and $15.0$ \% in Dice scores compared to the previous state-of-the-art. The proposed pipeline has the potential for application to a wide range of other tasks across various medical imaging modalities.
Efficient Pix2Vox++ for 3D Cardiac Reconstruction from 2D echo views
Jul 27, 2022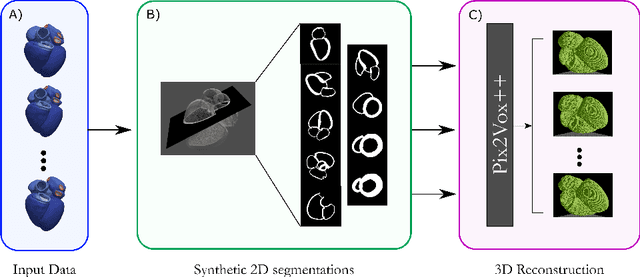
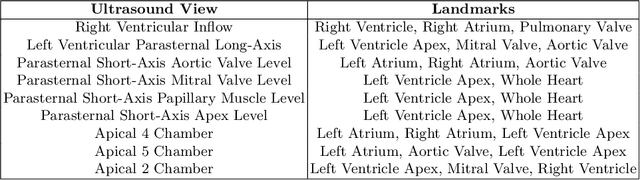
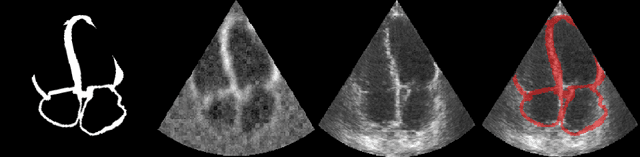
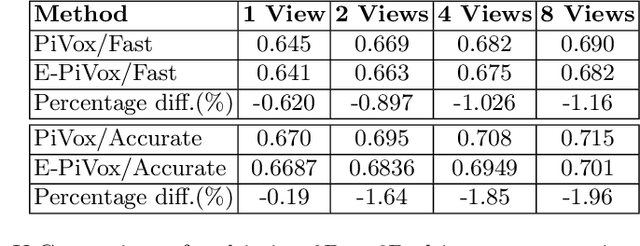
Abstract:Accurate geometric quantification of the human heart is a key step in the diagnosis of numerous cardiac diseases, and in the management of cardiac patients. Ultrasound imaging is the primary modality for cardiac imaging, however acquisition requires high operator skill, and its interpretation and analysis is difficult due to artifacts. Reconstructing cardiac anatomy in 3D can enable discovery of new biomarkers and make imaging less dependent on operator expertise, however most ultrasound systems only have 2D imaging capabilities. We propose both a simple alteration to the Pix2Vox++ networks for a sizeable reduction in memory usage and computational complexity, and a pipeline to perform reconstruction of 3D anatomy from 2D standard cardiac views, effectively enabling 3D anatomical reconstruction from limited 2D data. We evaluate our pipeline using synthetically generated data achieving accurate 3D whole-heart reconstructions (peak intersection over union score > 0.88) from just two standard anatomical 2D views of the heart. We also show preliminary results using real echo images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge