Matthew Sinclair
Scalable Agentic Reasoning for Designing Biologics Targeting Intrinsically Disordered Proteins
Dec 17, 2025Abstract:Intrinsically disordered proteins (IDPs) represent crucial therapeutic targets due to their significant role in disease -- approximately 80\% of cancer-related proteins contain long disordered regions -- but their lack of stable secondary/tertiary structures makes them "undruggable". While recent computational advances, such as diffusion models, can design high-affinity IDP binders, translating these to practical drug discovery requires autonomous systems capable of reasoning across complex conformational ensembles and orchestrating diverse computational tools at scale.To address this challenge, we designed and implemented StructBioReasoner, a scalable multi-agent system for designing biologics that can be used to target IDPs. StructBioReasoner employs a novel tournament-based reasoning framework where specialized agents compete to generate and refine therapeutic hypotheses, naturally distributing computational load for efficient exploration of the vast design space. Agents integrate domain knowledge with access to literature synthesis, AI-structure prediction, molecular simulations, and stability analysis, coordinating their execution on HPC infrastructure via an extensible federated agentic middleware, Academy. We benchmark StructBioReasoner across Der f 21 and NMNAT-2 and demonstrate that over 50\% of 787 designed and validated candidates for Der f 21 outperformed the human-designed reference binders from literature, in terms of improved binding free energy. For the more challenging NMNAT-2 protein, we identified three binding modes from 97,066 binders, including the well-studied NMNAT2:p53 interface. Thus, StructBioReasoner lays the groundwork for agentic reasoning systems for IDP therapeutic discovery on Exascale platforms.
Vector Representations of Vessel Trees
Jun 11, 2025Abstract:We introduce a novel framework for learning vector representations of tree-structured geometric data focusing on 3D vascular networks. Our approach employs two sequentially trained Transformer-based autoencoders. In the first stage, the Vessel Autoencoder captures continuous geometric details of individual vessel segments by learning embeddings from sampled points along each curve. In the second stage, the Vessel Tree Autoencoder encodes the topology of the vascular network as a single vector representation, leveraging the segment-level embeddings from the first model. A recursive decoding process ensures that the reconstructed topology is a valid tree structure. Compared to 3D convolutional models, this proposed approach substantially lowers GPU memory requirements, facilitating large-scale training. Experimental results on a 2D synthetic tree dataset and a 3D coronary artery dataset demonstrate superior reconstruction fidelity, accurate topology preservation, and realistic interpolations in latent space. Our scalable framework, named VeTTA, offers precise, flexible, and topologically consistent modeling of anatomical tree structures in medical imaging.
Improved 3D Whole Heart Geometry from Sparse CMR Slices
Aug 14, 2024Abstract:Cardiac magnetic resonance (CMR) imaging and computed tomography (CT) are two common non-invasive imaging methods for assessing patients with cardiovascular disease. CMR typically acquires multiple sparse 2D slices, with unavoidable respiratory motion artefacts between slices, whereas CT acquires isotropic dense data but uses ionising radiation. In this study, we explore the combination of Slice Shifting Algorithm (SSA), Spatial Transformer Network (STN), and Label Transformer Network (LTN) to: 1) correct respiratory motion between segmented slices, and 2) transform sparse segmentation data into dense segmentation. All combinations were validated using synthetic motion-corrupted CMR slice segmentation generated from CT in 1699 cases, where the dense CT serves as the ground truth. In 199 testing cases, SSA-LTN achieved the best results for Dice score and Huasdorff distance (94.0% and 4.7 mm respectively, average over 5 labels) but gave topological errors in 8 cases. STN was effective as a plug-in tool for correcting all topological errors with minimal impact on overall performance (93.5% and 5.0 mm respectively). SSA also proves to be a valuable plug-in tool, enhancing performance over both STN-based and LTN-based models. The code for these different combinations is available at https://github.com/XESchong/STACOM2024.
Pay Attention to the Atlas: Atlas-Guided Test-Time Adaptation Method for Robust 3D Medical Image Segmentation
Jul 02, 2023



Abstract:Convolutional neural networks (CNNs) often suffer from poor performance when tested on target data that differs from the training (source) data distribution, particularly in medical imaging applications where variations in imaging protocols across different clinical sites and scanners lead to different imaging appearances. However, re-accessing source training data for unsupervised domain adaptation or labeling additional test data for model fine-tuning can be difficult due to privacy issues and high labeling costs, respectively. To solve this problem, we propose a novel atlas-guided test-time adaptation (TTA) method for robust 3D medical image segmentation, called AdaAtlas. AdaAtlas only takes one single unlabeled test sample as input and adapts the segmentation network by minimizing an atlas-based loss. Specifically, the network is adapted so that its prediction after registration is aligned with the learned atlas in the atlas space, which helps to reduce anatomical segmentation errors at test time. In addition, different from most existing TTA methods which restrict the adaptation to batch normalization blocks in the segmentation network only, we further exploit the use of channel and spatial attention blocks for improved adaptability at test time. Extensive experiments on multiple datasets from different sites show that AdaAtlas with attention blocks adapted (AdaAtlas-Attention) achieves superior performance improvements, greatly outperforming other competitive TTA methods.
Image To Tree with Recursive Prompting
Jan 01, 2023



Abstract:Extracting complex structures from grid-based data is a common key step in automated medical image analysis. The conventional solution to recovering tree-structured geometries typically involves computing the minimal cost path through intermediate representations derived from segmentation masks. However, this methodology has significant limitations in the context of projective imaging of tree-structured 3D anatomical data such as coronary arteries, since there are often overlapping branches in the 2D projection. In this work, we propose a novel approach to predicting tree connectivity structure which reformulates the task as an optimization problem over individual steps of a recursive process. We design and train a two-stage model which leverages the UNet and Transformer architectures and introduces an image-based prompting technique. Our proposed method achieves compelling results on a pair of synthetic datasets, and outperforms a shortest-path baseline.
CAS-Net: Conditional Atlas Generation and Brain Segmentation for Fetal MRI
May 17, 2022Abstract:Fetal Magnetic Resonance Imaging (MRI) is used in prenatal diagnosis and to assess early brain development. Accurate segmentation of the different brain tissues is a vital step in several brain analysis tasks, such as cortical surface reconstruction and tissue thickness measurements. Fetal MRI scans, however, are prone to motion artifacts that can affect the correctness of both manual and automatic segmentation techniques. In this paper, we propose a novel network structure that can simultaneously generate conditional atlases and predict brain tissue segmentation, called CAS-Net. The conditional atlases provide anatomical priors that can constrain the segmentation connectivity, despite the heterogeneity of intensity values caused by motion or partial volume effects. The proposed method is trained and evaluated on 253 subjects from the developing Human Connectome Project (dHCP). The results demonstrate that the proposed method can generate conditional age-specific atlas with sharp boundary and shape variance. It also segment multi-category brain tissues for fetal MRI with a high overall Dice similarity coefficient (DSC) of $85.2\%$ for the selected 9 tissue labels.
Detecting Hypo-plastic Left Heart Syndrome in Fetal Ultrasound via Disease-specific Atlas Maps
Jul 06, 2021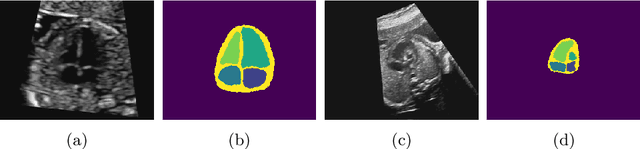
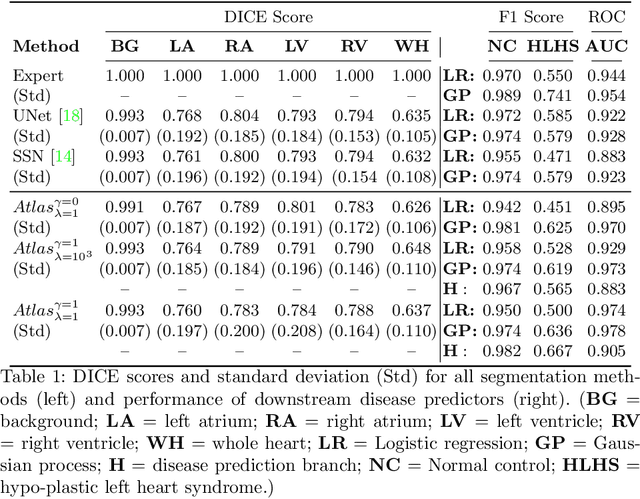
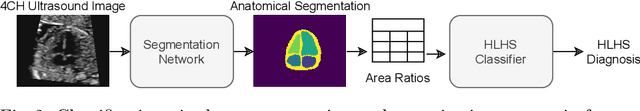
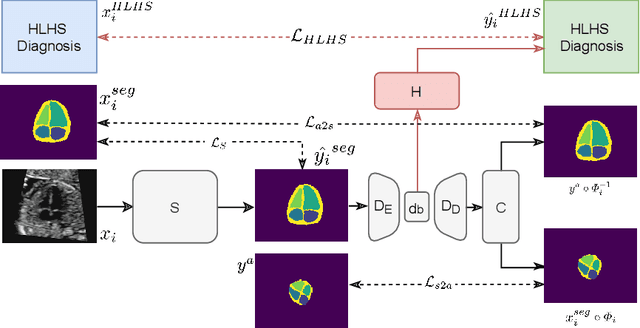
Abstract:Fetal ultrasound screening during pregnancy plays a vital role in the early detection of fetal malformations which have potential long-term health impacts. The level of skill required to diagnose such malformations from live ultrasound during examination is high and resources for screening are often limited. We present an interpretable, atlas-learning segmentation method for automatic diagnosis of Hypo-plastic Left Heart Syndrome (HLHS) from a single `4 Chamber Heart' view image. We propose to extend the recently introduced Image-and-Spatial Transformer Networks (Atlas-ISTN) into a framework that enables sensitising atlas generation to disease. In this framework we can jointly learn image segmentation, registration, atlas construction and disease prediction while providing a maximum level of clinical interpretability compared to direct image classification methods. As a result our segmentation allows diagnoses competitive with expert-derived manual diagnosis and yields an AUC-ROC of 0.978 (1043 cases for training, 260 for validation and 325 for testing).
Atlas-ISTN: Joint Segmentation, Registration and Atlas Construction with Image-and-Spatial Transformer Networks
Dec 18, 2020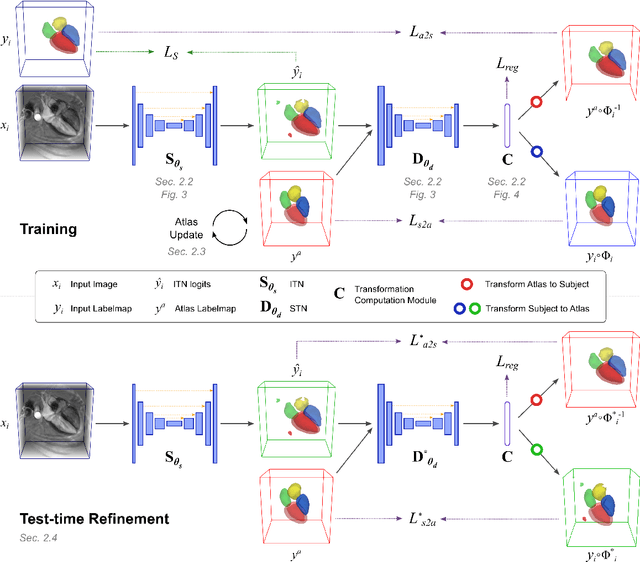
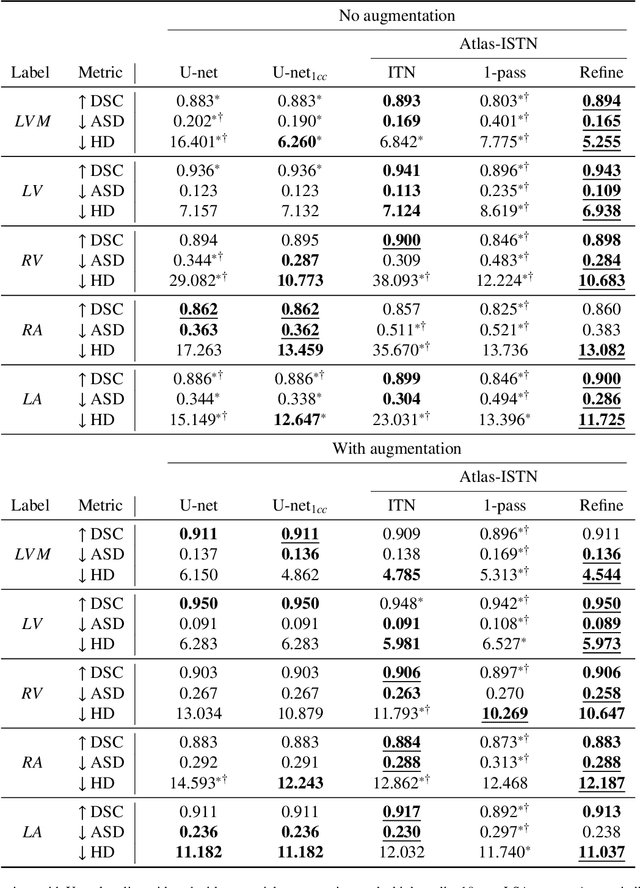
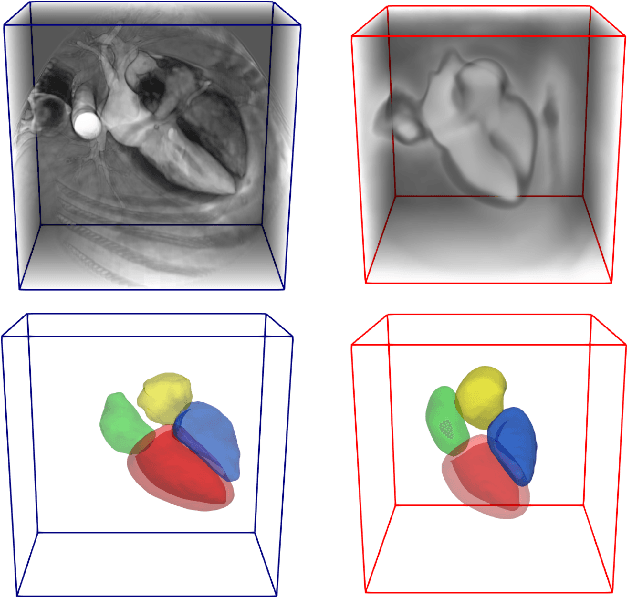
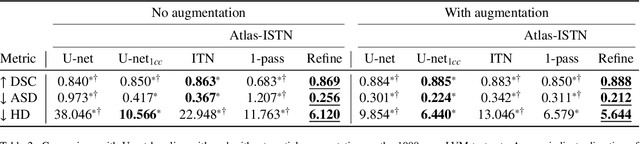
Abstract:Deep learning models for semantic segmentation are able to learn powerful representations for pixel-wise predictions, but are sensitive to noise at test time and do not guarantee a plausible topology. Image registration models on the other hand are able to warp known topologies to target images as a means of segmentation, but typically require large amounts of training data, and have not widely been benchmarked against pixel-wise segmentation models. We propose Atlas-ISTN, a framework that jointly learns segmentation and registration on 2D and 3D image data, and constructs a population-derived atlas in the process. Atlas-ISTN learns to segment multiple structures of interest and to register the constructed, topologically consistent atlas labelmap to an intermediate pixel-wise segmentation. Additionally, Atlas-ISTN allows for test time refinement of the model's parameters to optimize the alignment of the atlas labelmap to an intermediate pixel-wise segmentation. This process both mitigates for noise in the target image that can result in spurious pixel-wise predictions, as well as improves upon the one-pass prediction of the model. Benefits of the Atlas-ISTN framework are demonstrated qualitatively and quantitatively on 2D synthetic data and 3D cardiac computed tomography and brain magnetic resonance image data, out-performing both segmentation and registration baseline models. Atlas-ISTN also provides inter-subject correspondence of the structures of interest, enabling population-level shape and motion analysis.
Automated quantification of myocardial tissue characteristics from native T1 mapping using neural networks with Bayesian inference for uncertainty-based quality-control
Jan 31, 2020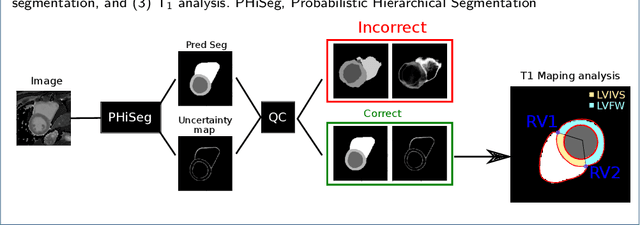
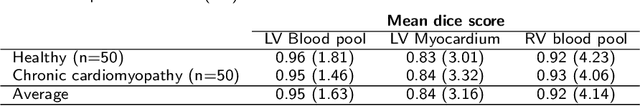
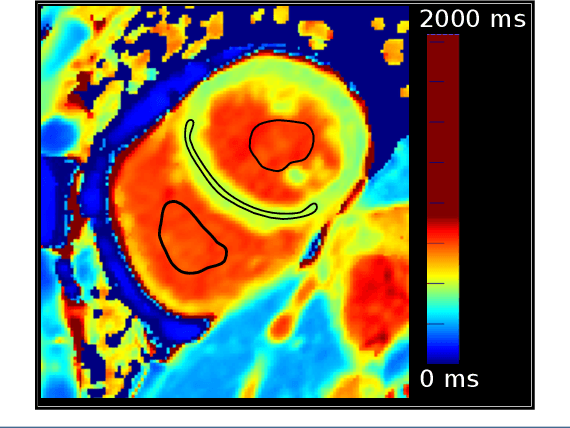

Abstract:Tissue characterisation with CMR parametric mapping has the potential to detect and quantify both focal and diffuse alterations in myocardial structure not assessable by late gadolinium enhancement. Native T1 mapping in particular has shown promise as a useful biomarker to support diagnostic, therapeutic and prognostic decision-making in ischaemic and non-ischaemic cardiomyopathies. Convolutional neural networks with Bayesian inference are a category of artificial neural networks which model the uncertainty of the network output. This study presents an automated framework for tissue characterisation from native ShMOLLI T1 mapping at 1.5T using a Probabilistic Hierarchical Segmentation (PHiSeg) network. In addition, we use the uncertainty information provided by the PHiSeg network in a novel automated quality control (QC) step to identify uncertain T1 values. The PHiSeg network and QC were validated against manual analysis on a cohort of the UK Biobank containing healthy subjects and chronic cardiomyopathy patients. We used the proposed method to obtain reference T1 ranges for the left ventricular myocardium in healthy subjects as well as common clinical cardiac conditions. T1 values computed from automatic and manual segmentations were highly correlated (r=0.97). Bland-Altman analysis showed good agreement between the automated and manual measurements. The average Dice metric was 0.84 for the left ventricular myocardium. The sensitivity of detection of erroneous outputs was 91%. Finally, T1 values were automatically derived from 14,683 CMR exams from the UK Biobank. The proposed pipeline allows for automatic analysis of myocardial native T1 mapping and includes a QC process to detect potentially erroneous results. T1 reference values were presented for healthy subjects and common clinical cardiac conditions from the largest cohort to date using T1-mapping images.
Confident Head Circumference Measurement from Ultrasound with Real-time Feedback for Sonographers
Aug 07, 2019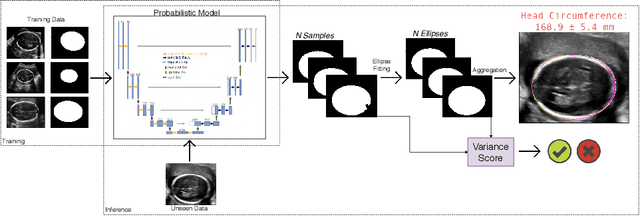
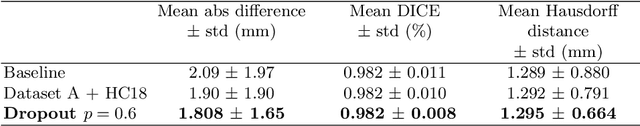
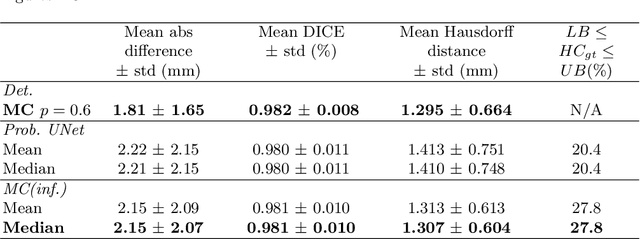
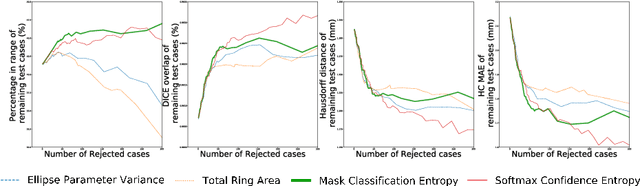
Abstract:Manual estimation of fetal Head Circumference (HC) from Ultrasound (US) is a key biometric for monitoring the healthy development of fetuses. Unfortunately, such measurements are subject to large inter-observer variability, resulting in low early-detection rates of fetal abnormalities. To address this issue, we propose a novel probabilistic Deep Learning approach for real-time automated estimation of fetal HC. This system feeds back statistics on measurement robustness to inform users how confident a deep neural network is in evaluating suitable views acquired during free-hand ultrasound examination. In real-time scenarios, this approach may be exploited to guide operators to scan planes that are as close as possible to the underlying distribution of training images, for the purpose of improving inter-operator consistency. We train on free-hand ultrasound data from over 2000 subjects (2848 training/540 test) and show that our method is able to predict HC measurements within 1.81$\pm$1.65mm deviation from the ground truth, with 50% of the test images fully contained within the predicted confidence margins, and an average of 1.82$\pm$1.78mm deviation from the margin for the remaining cases that are not fully contained.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge