Thomas Day
EchoNet-Synthetic: Privacy-preserving Video Generation for Safe Medical Data Sharing
Jun 02, 2024Abstract:To make medical datasets accessible without sharing sensitive patient information, we introduce a novel end-to-end approach for generative de-identification of dynamic medical imaging data. Until now, generative methods have faced constraints in terms of fidelity, spatio-temporal coherence, and the length of generation, failing to capture the complete details of dataset distributions. We present a model designed to produce high-fidelity, long and complete data samples with near-real-time efficiency and explore our approach on a challenging task: generating echocardiogram videos. We develop our generation method based on diffusion models and introduce a protocol for medical video dataset anonymization. As an exemplar, we present EchoNet-Synthetic, a fully synthetic, privacy-compliant echocardiogram dataset with paired ejection fraction labels. As part of our de-identification protocol, we evaluate the quality of the generated dataset and propose to use clinical downstream tasks as a measurement on top of widely used but potentially biased image quality metrics. Experimental outcomes demonstrate that EchoNet-Synthetic achieves comparable dataset fidelity to the actual dataset, effectively supporting the ejection fraction regression task. Code, weights and dataset are available at https://github.com/HReynaud/EchoNet-Synthetic.
Feature-Conditioned Cascaded Video Diffusion Models for Precise Echocardiogram Synthesis
Mar 23, 2023Abstract:Image synthesis is expected to provide value for the translation of machine learning methods into clinical practice. Fundamental problems like model robustness, domain transfer, causal modelling, and operator training become approachable through synthetic data. Especially, heavily operator-dependant modalities like Ultrasound imaging require robust frameworks for image and video generation. So far, video generation has only been possible by providing input data that is as rich as the output data, e.g., image sequence plus conditioning in, video out. However, clinical documentation is usually scarce and only single images are reported and stored, thus retrospective patient-specific analysis or the generation of rich training data becomes impossible with current approaches. In this paper, we extend elucidated diffusion models for video modelling to generate plausible video sequences from single images and arbitrary conditioning with clinical parameters. We explore this idea within the context of echocardiograms by looking into the variation of the Left Ventricle Ejection Fraction, the most essential clinical metric gained from these examinations. We use the publicly available EchoNet-Dynamic dataset for all our experiments. Our image to sequence approach achieves an $R^2$ score of 93%, which is 38 points higher than recently proposed sequence to sequence generation methods. Code and models will be available at: https://github.com/HReynaud/EchoDiffusion.
Can non-specialists provide high quality gold standard labels in challenging modalities?
Jul 30, 2021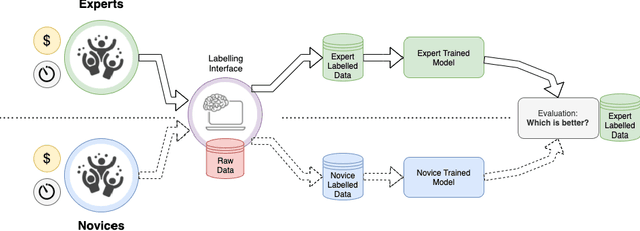
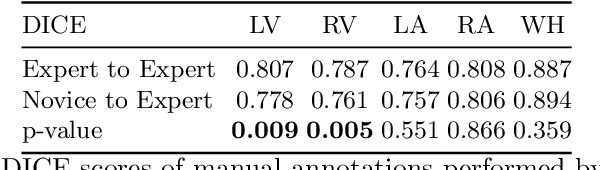
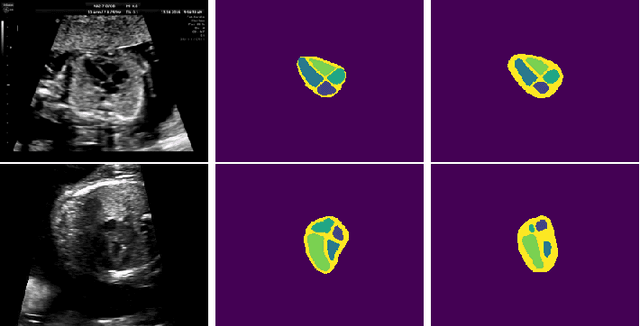
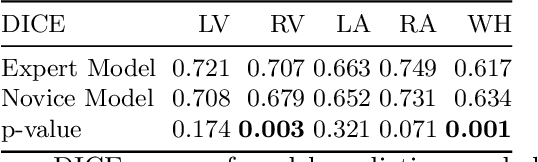
Abstract:Probably yes. -- Supervised Deep Learning dominates performance scores for many computer vision tasks and defines the state-of-the-art. However, medical image analysis lags behind natural image applications. One of the many reasons is the lack of well annotated medical image data available to researchers. One of the first things researchers are told is that we require significant expertise to reliably and accurately interpret and label such data. We see significant inter- and intra-observer variability between expert annotations of medical images. Still, it is a widely held assumption that novice annotators are unable to provide useful annotations for use by clinical Deep Learning models. In this work we challenge this assumption and examine the implications of using a minimally trained novice labelling workforce to acquire annotations for a complex medical image dataset. We study the time and cost implications of using novice annotators, the raw performance of novice annotators compared to gold-standard expert annotators, and the downstream effects on a trained Deep Learning segmentation model's performance for detecting a specific congenital heart disease (hypoplastic left heart syndrome) in fetal ultrasound imaging.
Detecting Hypo-plastic Left Heart Syndrome in Fetal Ultrasound via Disease-specific Atlas Maps
Jul 06, 2021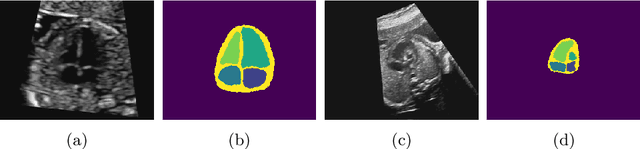
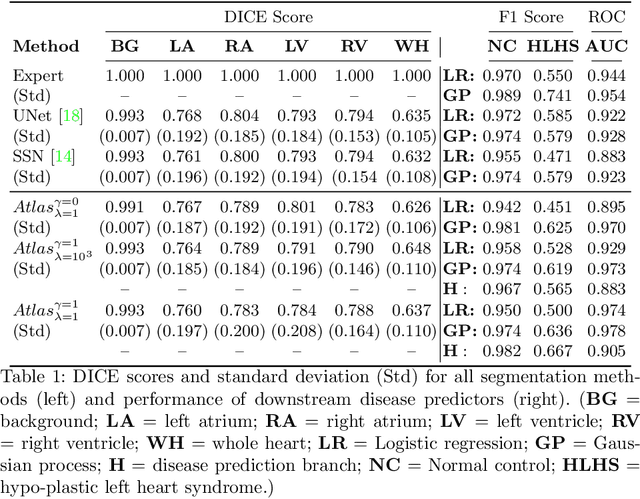
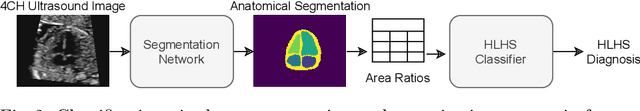
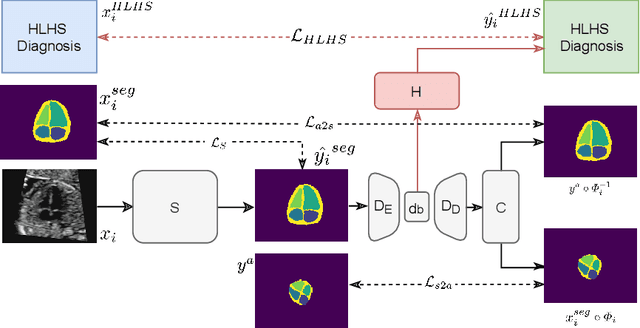
Abstract:Fetal ultrasound screening during pregnancy plays a vital role in the early detection of fetal malformations which have potential long-term health impacts. The level of skill required to diagnose such malformations from live ultrasound during examination is high and resources for screening are often limited. We present an interpretable, atlas-learning segmentation method for automatic diagnosis of Hypo-plastic Left Heart Syndrome (HLHS) from a single `4 Chamber Heart' view image. We propose to extend the recently introduced Image-and-Spatial Transformer Networks (Atlas-ISTN) into a framework that enables sensitising atlas generation to disease. In this framework we can jointly learn image segmentation, registration, atlas construction and disease prediction while providing a maximum level of clinical interpretability compared to direct image classification methods. As a result our segmentation allows diagnoses competitive with expert-derived manual diagnosis and yields an AUC-ROC of 0.978 (1043 cases for training, 260 for validation and 325 for testing).
Detecting Outliers with Poisson Image Interpolation
Jul 06, 2021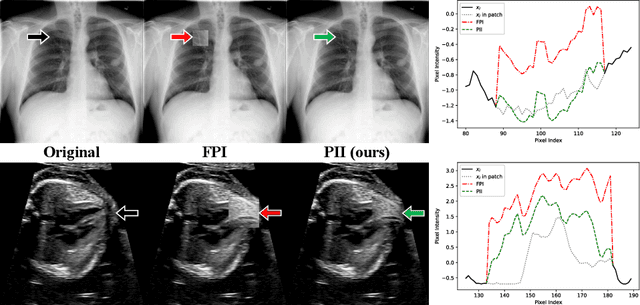
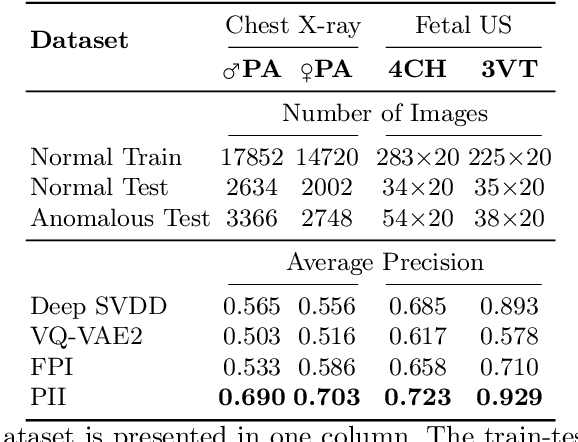
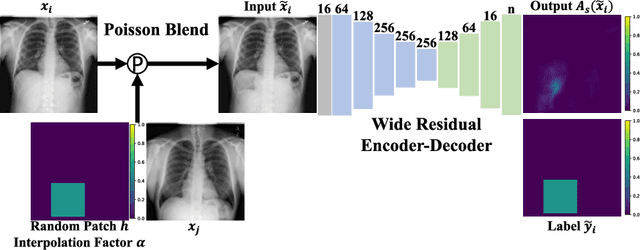
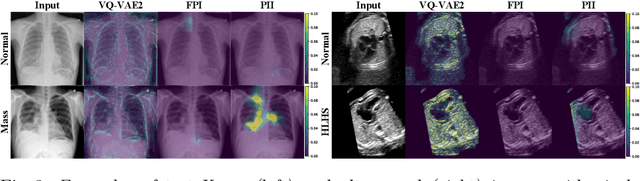
Abstract:Supervised learning of every possible pathology is unrealistic for many primary care applications like health screening. Image anomaly detection methods that learn normal appearance from only healthy data have shown promising results recently. We propose an alternative to image reconstruction-based and image embedding-based methods and propose a new self-supervised method to tackle pathological anomaly detection. Our approach originates in the foreign patch interpolation (FPI) strategy that has shown superior performance on brain MRI and abdominal CT data. We propose to use a better patch interpolation strategy, Poisson image interpolation (PII), which makes our method suitable for applications in challenging data regimes. PII outperforms state-of-the-art methods by a good margin when tested on surrogate tasks like identifying common lung anomalies in chest X-rays or hypo-plastic left heart syndrome in prenatal, fetal cardiac ultrasound images. Code available at https://github.com/jemtan/PII.
Learning normal appearance for fetal anomaly screening: Application to the unsupervised detection of Hypoplastic Left Heart Syndrome
Nov 15, 2020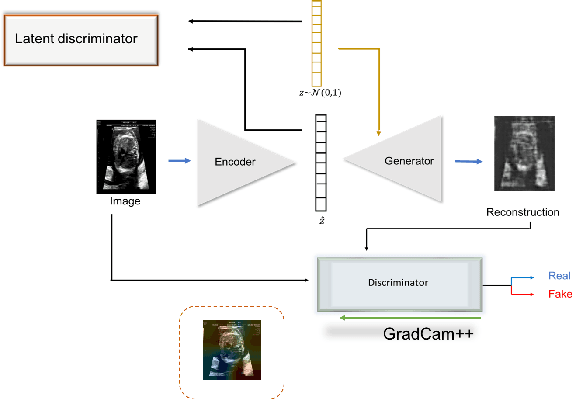
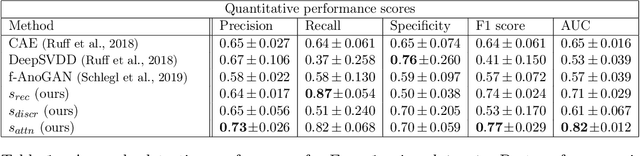
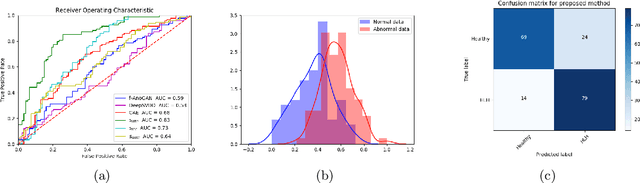
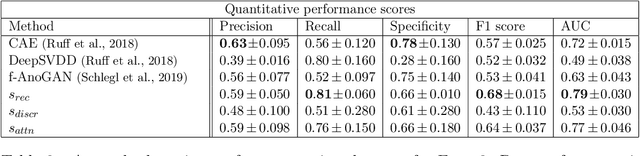
Abstract:Congenital heart disease is considered as one the most common groups of congenital malformations which affects $6-11$ per $1000$ newborns. In this work, an automated framework for detection of cardiac anomalies during ultrasound screening is proposed and evaluated on the example of Hypoplastic Left Heart Syndrome (HLHS), a sub-category of congenital heart disease. We propose an unsupervised approach that learns healthy anatomy exclusively from clinically confirmed normal control patients. We evaluate a number of known anomaly detection frameworks together with a new model architecture based on the $\alpha$-GAN network and find evidence that the proposed model performs significantly better than the state-of-the-art in image-based anomaly detection, yielding average $0.81$ AUC \emph{and} a better robustness towards initialisation compared to previous works.
Automated Detection of Congenital Heart Disease in Fetal Ultrasound Screening
Aug 18, 2020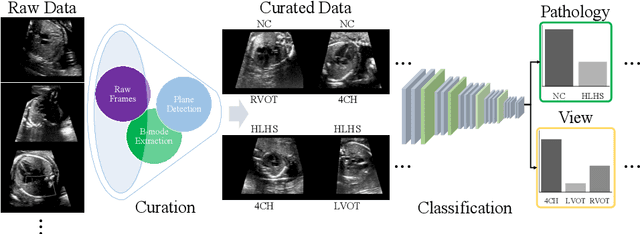
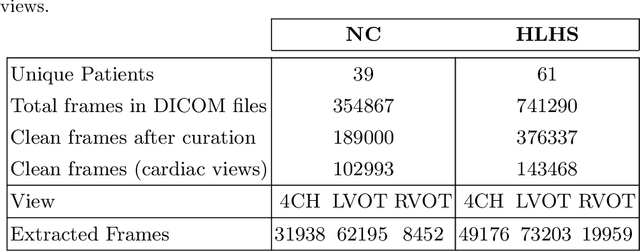
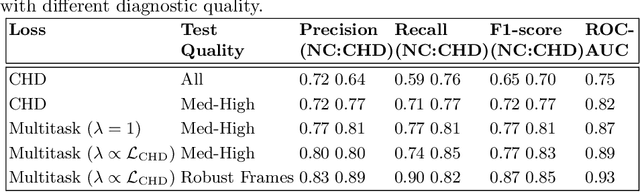
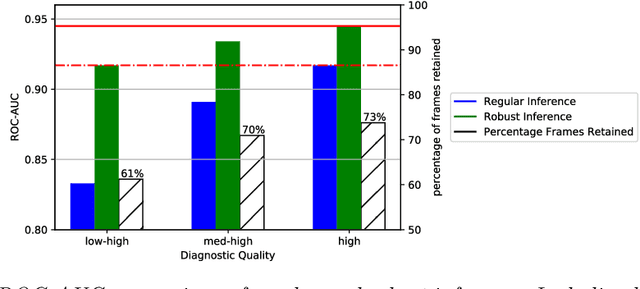
Abstract:Prenatal screening with ultrasound can lower neonatal mortality significantly for selected cardiac abnormalities. However, the need for human expertise, coupled with the high volume of screening cases, limits the practically achievable detection rates. In this paper we discuss the potential for deep learning techniques to aid in the detection of congenital heart disease (CHD) in fetal ultrasound. We propose a pipeline for automated data curation and classification. During both training and inference, we exploit an auxiliary view classification task to bias features toward relevant cardiac structures. This bias helps to improve in F1-scores from 0.72 and 0.77 to 0.87 and 0.85 for healthy and CHD classes respectively.
Recent Developments and Future Challenges in Medical Mixed Reality
Aug 03, 2017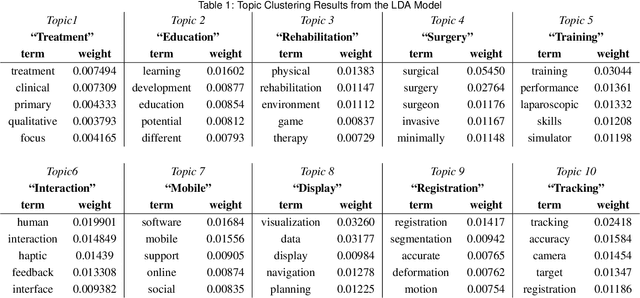
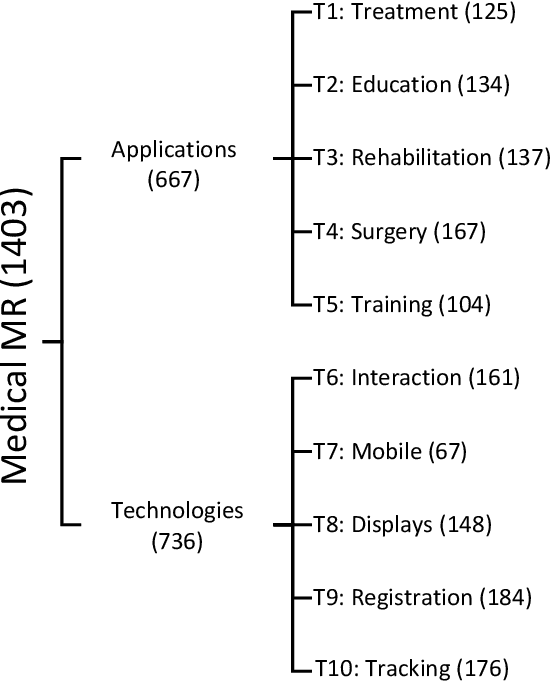
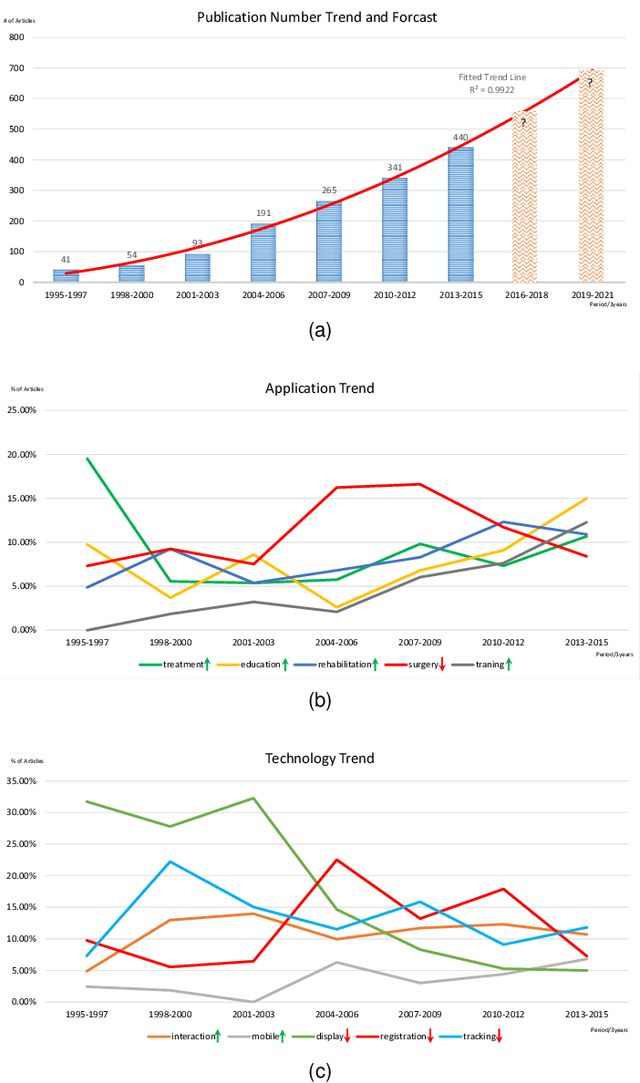
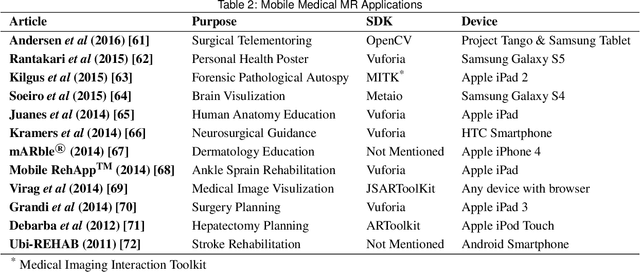
Abstract:Mixed Reality (MR) is of increasing interest within technology-driven modern medicine but is not yet used in everyday practice. This situation is changing rapidly, however, and this paper explores the emergence of MR technology and the importance of its utility within medical applications. A classification of medical MR has been obtained by applying an unbiased text mining method to a database of 1,403 relevant research papers published over the last two decades. The classification results reveal a taxonomy for the development of medical MR research during this period as well as suggesting future trends. We then use the classification to analyse the technology and applications developed in the last five years. Our objective is to aid researchers to focus on the areas where technology advancements in medical MR are most needed, as well as providing medical practitioners with a useful source of reference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge