Qingjie Meng
MCM: Mamba-based Cardiac Motion Tracking using Sequential Images in MRI
Jul 23, 2025Abstract:Myocardial motion tracking is important for assessing cardiac function and diagnosing cardiovascular diseases, for which cine cardiac magnetic resonance (CMR) has been established as the gold standard imaging modality. Many existing methods learn motion from single image pairs consisting of a reference frame and a randomly selected target frame from the cardiac cycle. However, these methods overlook the continuous nature of cardiac motion and often yield inconsistent and non-smooth motion estimations. In this work, we propose a novel Mamba-based cardiac motion tracking network (MCM) that explicitly incorporates target image sequence from the cardiac cycle to achieve smooth and temporally consistent motion tracking. By developing a bi-directional Mamba block equipped with a bi-directional scanning mechanism, our method facilitates the estimation of plausible deformation fields. With our proposed motion decoder that integrates motion information from frames adjacent to the target frame, our method further enhances temporal coherence. Moreover, by taking advantage of Mamba's structured state-space formulation, the proposed method learns the continuous dynamics of the myocardium from sequential images without increasing computational complexity. We evaluate the proposed method on two public datasets. The experimental results demonstrate that the proposed method quantitatively and qualitatively outperforms both conventional and state-of-the-art learning-based cardiac motion tracking methods. The code is available at https://github.com/yjh-0104/MCM.
EchoFlow: A Foundation Model for Cardiac Ultrasound Image and Video Generation
Mar 28, 2025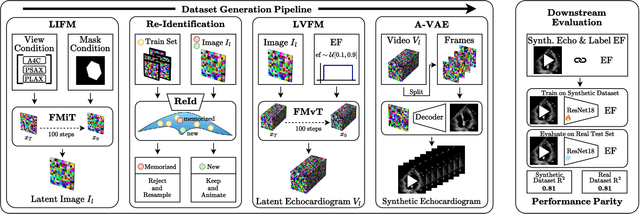
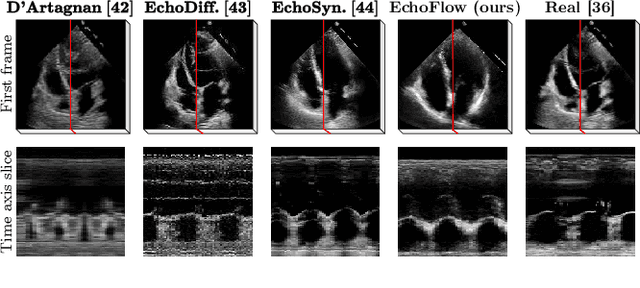
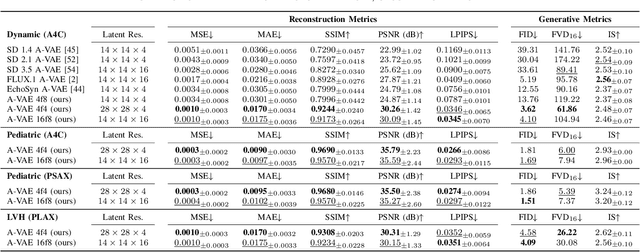
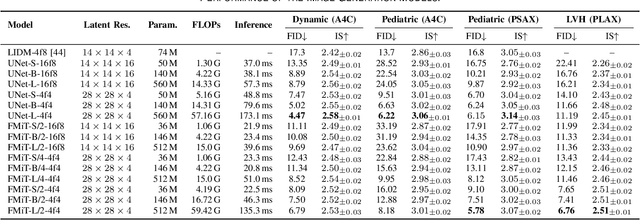
Abstract:Advances in deep learning have significantly enhanced medical image analysis, yet the availability of large-scale medical datasets remains constrained by patient privacy concerns. We present EchoFlow, a novel framework designed to generate high-quality, privacy-preserving synthetic echocardiogram images and videos. EchoFlow comprises four key components: an adversarial variational autoencoder for defining an efficient latent representation of cardiac ultrasound images, a latent image flow matching model for generating accurate latent echocardiogram images, a latent re-identification model to ensure privacy by filtering images anatomically, and a latent video flow matching model for animating latent images into realistic echocardiogram videos conditioned on ejection fraction. We rigorously evaluate our synthetic datasets on the clinically relevant task of ejection fraction regression and demonstrate, for the first time, that downstream models trained exclusively on EchoFlow-generated synthetic datasets achieve performance parity with models trained on real datasets. We release our models and synthetic datasets, enabling broader, privacy-compliant research in medical ultrasound imaging at https://huggingface.co/spaces/HReynaud/EchoFlow.
SACB-Net: Spatial-awareness Convolutions for Medical Image Registration
Mar 25, 2025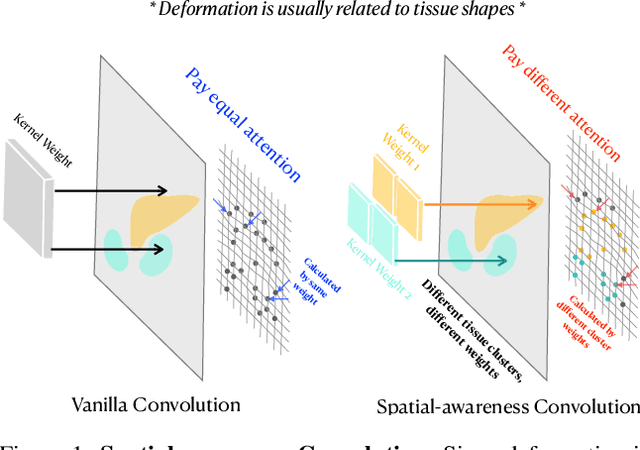
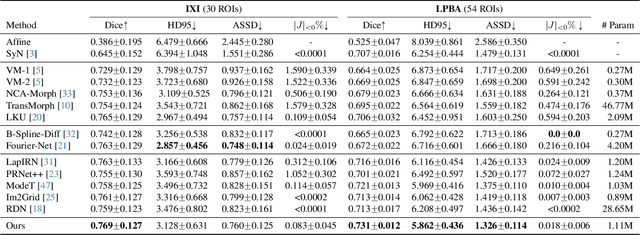
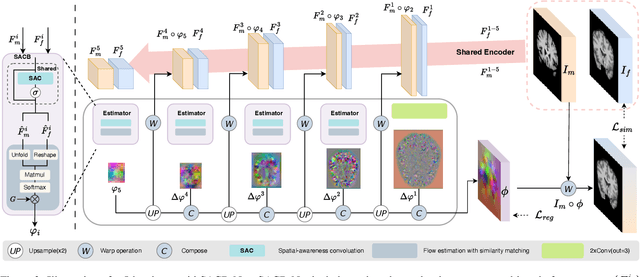
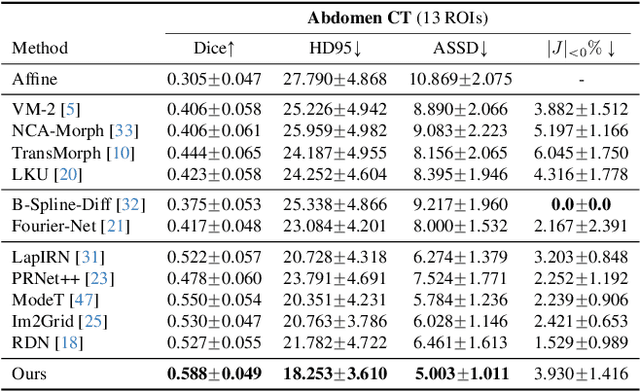
Abstract:Deep learning-based image registration methods have shown state-of-the-art performance and rapid inference speeds. Despite these advances, many existing approaches fall short in capturing spatially varying information in non-local regions of feature maps due to the reliance on spatially-shared convolution kernels. This limitation leads to suboptimal estimation of deformation fields. In this paper, we propose a 3D Spatial-Awareness Convolution Block (SACB) to enhance the spatial information within feature representations. Our SACB estimates the spatial clusters within feature maps by leveraging feature similarity and subsequently parameterizes the adaptive convolution kernels across diverse regions. This adaptive mechanism generates the convolution kernels (weights and biases) tailored to spatial variations, thereby enabling the network to effectively capture spatially varying information. Building on SACB, we introduce a pyramid flow estimator (named SACB-Net) that integrates SACBs to facilitate multi-scale flow composition, particularly addressing large deformations. Experimental results on the brain IXI and LPBA datasets as well as Abdomen CT datasets demonstrate the effectiveness of SACB and the superiority of SACB-Net over the state-of-the-art learning-based registration methods. The code is available at https://github.com/x-xc/SACB_Net .
JVID: Joint Video-Image Diffusion for Visual-Quality and Temporal-Consistency in Video Generation
Sep 21, 2024



Abstract:We introduce the Joint Video-Image Diffusion model (JVID), a novel approach to generating high-quality and temporally coherent videos. We achieve this by integrating two diffusion models: a Latent Image Diffusion Model (LIDM) trained on images and a Latent Video Diffusion Model (LVDM) trained on video data. Our method combines these models in the reverse diffusion process, where the LIDM enhances image quality and the LVDM ensures temporal consistency. This unique combination allows us to effectively handle the complex spatio-temporal dynamics in video generation. Our results demonstrate quantitative and qualitative improvements in producing realistic and coherent videos.
EchoNet-Synthetic: Privacy-preserving Video Generation for Safe Medical Data Sharing
Jun 02, 2024Abstract:To make medical datasets accessible without sharing sensitive patient information, we introduce a novel end-to-end approach for generative de-identification of dynamic medical imaging data. Until now, generative methods have faced constraints in terms of fidelity, spatio-temporal coherence, and the length of generation, failing to capture the complete details of dataset distributions. We present a model designed to produce high-fidelity, long and complete data samples with near-real-time efficiency and explore our approach on a challenging task: generating echocardiogram videos. We develop our generation method based on diffusion models and introduce a protocol for medical video dataset anonymization. As an exemplar, we present EchoNet-Synthetic, a fully synthetic, privacy-compliant echocardiogram dataset with paired ejection fraction labels. As part of our de-identification protocol, we evaluate the quality of the generated dataset and propose to use clinical downstream tasks as a measurement on top of widely used but potentially biased image quality metrics. Experimental outcomes demonstrate that EchoNet-Synthetic achieves comparable dataset fidelity to the actual dataset, effectively supporting the ejection fraction regression task. Code, weights and dataset are available at https://github.com/HReynaud/EchoNet-Synthetic.
DeepMesh: Mesh-based Cardiac Motion Tracking using Deep Learning
Sep 25, 2023



Abstract:3D motion estimation from cine cardiac magnetic resonance (CMR) images is important for the assessment of cardiac function and the diagnosis of cardiovascular diseases. Current state-of-the art methods focus on estimating dense pixel-/voxel-wise motion fields in image space, which ignores the fact that motion estimation is only relevant and useful within the anatomical objects of interest, e.g., the heart. In this work, we model the heart as a 3D mesh consisting of epi- and endocardial surfaces. We propose a novel learning framework, DeepMesh, which propagates a template heart mesh to a subject space and estimates the 3D motion of the heart mesh from CMR images for individual subjects. In DeepMesh, the heart mesh of the end-diastolic frame of an individual subject is first reconstructed from the template mesh. Mesh-based 3D motion fields with respect to the end-diastolic frame are then estimated from 2D short- and long-axis CMR images. By developing a differentiable mesh-to-image rasterizer, DeepMesh is able to leverage 2D shape information from multiple anatomical views for 3D mesh reconstruction and mesh motion estimation. The proposed method estimates vertex-wise displacement and thus maintains vertex correspondences between time frames, which is important for the quantitative assessment of cardiac function across different subjects and populations. We evaluate DeepMesh on CMR images acquired from the UK Biobank. We focus on 3D motion estimation of the left ventricle in this work. Experimental results show that the proposed method quantitatively and qualitatively outperforms other image-based and mesh-based cardiac motion tracking methods.
Mesh-based 3D Motion Tracking in Cardiac MRI using Deep Learning
Sep 05, 2022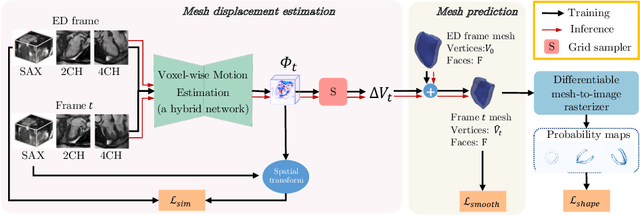
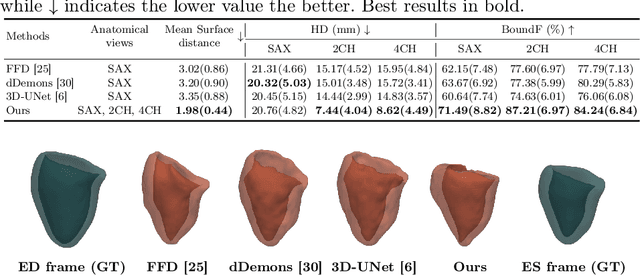
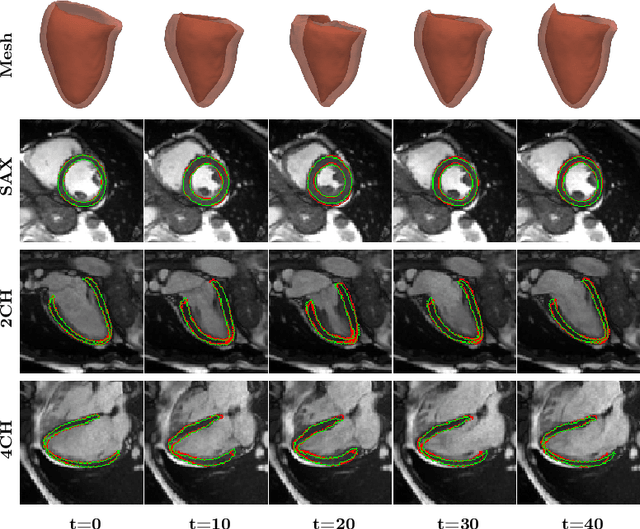

Abstract:3D motion estimation from cine cardiac magnetic resonance (CMR) images is important for the assessment of cardiac function and diagnosis of cardiovascular diseases. Most of the previous methods focus on estimating pixel-/voxel-wise motion fields in the full image space, which ignore the fact that motion estimation is mainly relevant and useful within the object of interest, e.g., the heart. In this work, we model the heart as a 3D geometric mesh and propose a novel deep learning-based method that can estimate 3D motion of the heart mesh from 2D short- and long-axis CMR images. By developing a differentiable mesh-to-image rasterizer, the method is able to leverage the anatomical shape information from 2D multi-view CMR images for 3D motion estimation. The differentiability of the rasterizer enables us to train the method end-to-end. One advantage of the proposed method is that by tracking the motion of each vertex, it is able to keep the vertex correspondence of 3D meshes between time frames, which is important for quantitative assessment of the cardiac function on the mesh. We evaluate the proposed method on CMR images acquired from the UK Biobank study. Experimental results show that the proposed method quantitatively and qualitatively outperforms both conventional and learning-based cardiac motion tracking methods.
MulViMotion: Shape-aware 3D Myocardial Motion Tracking from Multi-View Cardiac MRI
Jul 29, 2022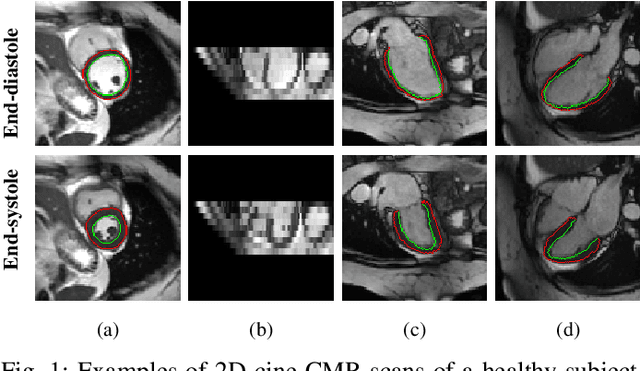
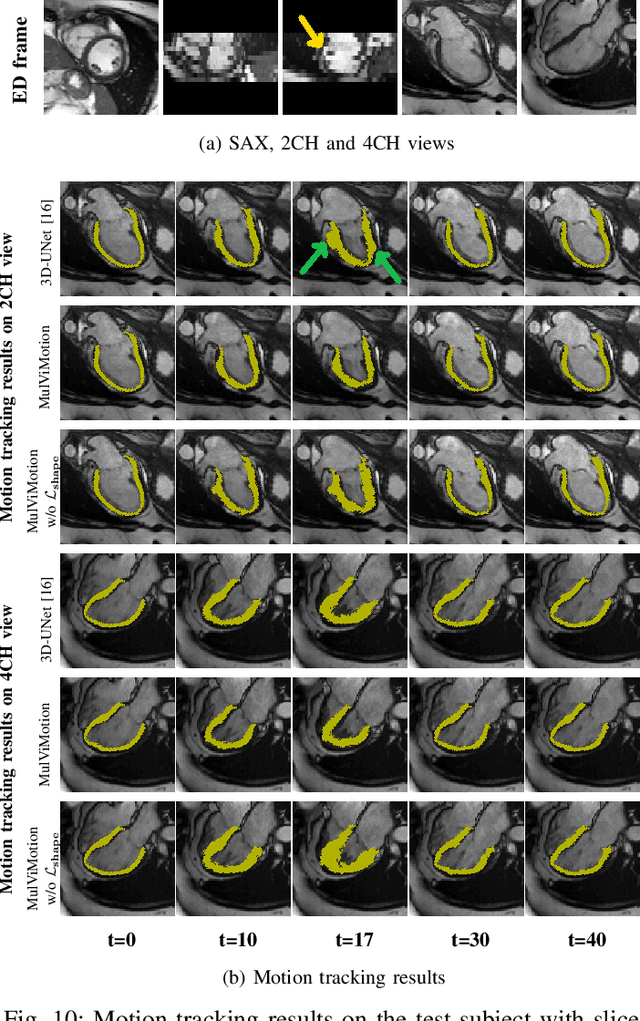
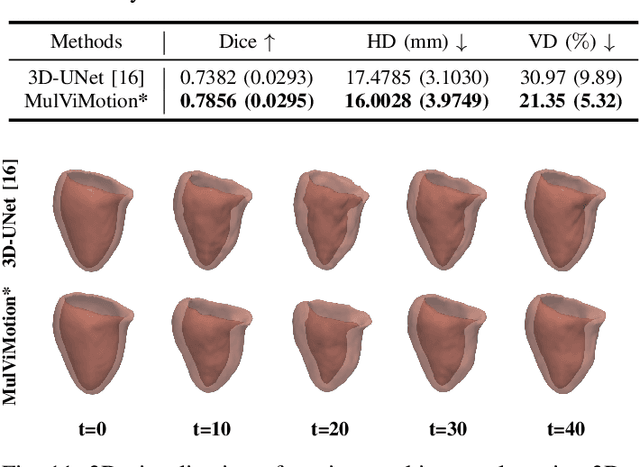
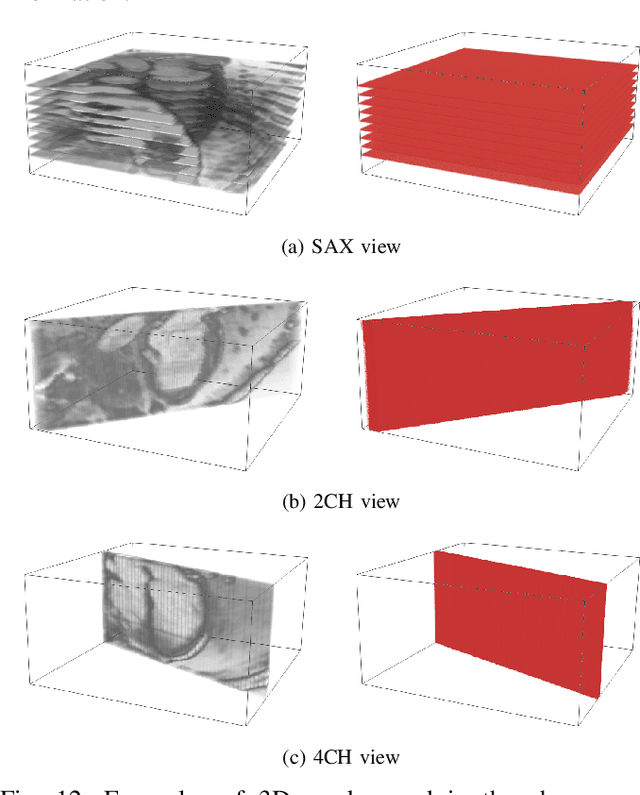
Abstract:Recovering the 3D motion of the heart from cine cardiac magnetic resonance (CMR) imaging enables the assessment of regional myocardial function and is important for understanding and analyzing cardiovascular disease. However, 3D cardiac motion estimation is challenging because the acquired cine CMR images are usually 2D slices which limit the accurate estimation of through-plane motion. To address this problem, we propose a novel multi-view motion estimation network (MulViMotion), which integrates 2D cine CMR images acquired in short-axis and long-axis planes to learn a consistent 3D motion field of the heart. In the proposed method, a hybrid 2D/3D network is built to generate dense 3D motion fields by learning fused representations from multi-view images. To ensure that the motion estimation is consistent in 3D, a shape regularization module is introduced during training, where shape information from multi-view images is exploited to provide weak supervision to 3D motion estimation. We extensively evaluate the proposed method on 2D cine CMR images from 580 subjects of the UK Biobank study for 3D motion tracking of the left ventricular myocardium. Experimental results show that the proposed method quantitatively and qualitatively outperforms competing methods.
Video Summarization through Reinforcement Learning with a 3D Spatio-Temporal U-Net
Jun 19, 2021
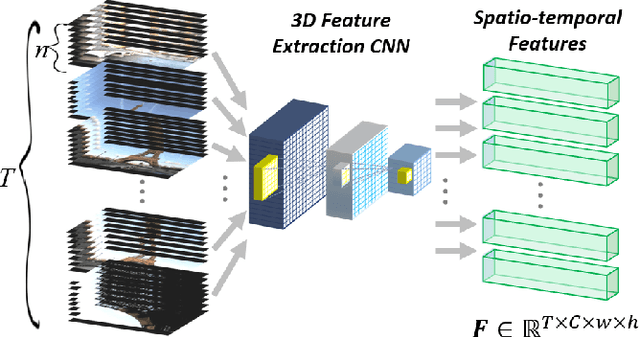
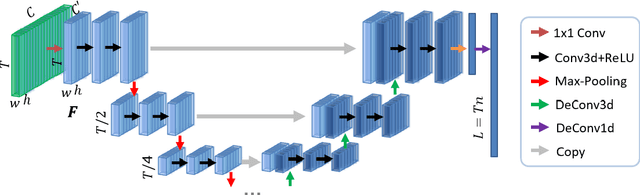
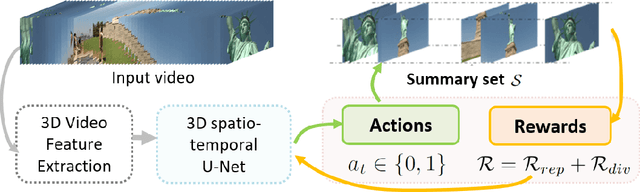
Abstract:Intelligent video summarization algorithms allow to quickly convey the most relevant information in videos through the identification of the most essential and explanatory content while removing redundant video frames. In this paper, we introduce the 3DST-UNet-RL framework for video summarization. A 3D spatio-temporal U-Net is used to efficiently encode spatio-temporal information of the input videos for downstream reinforcement learning (RL). An RL agent learns from spatio-temporal latent scores and predicts actions for keeping or rejecting a video frame in a video summary. We investigate if real/inflated 3D spatio-temporal CNN features are better suited to learn representations from videos than commonly used 2D image features. Our framework can operate in both, a fully unsupervised mode and a supervised training mode. We analyse the impact of prescribed summary lengths and show experimental evidence for the effectiveness of 3DST-UNet-RL on two commonly used general video summarization benchmarks. We also applied our method on a medical video summarization task. The proposed video summarization method has the potential to save storage costs of ultrasound screening videos as well as to increase efficiency when browsing patient video data during retrospective analysis or audit without loosing essential information
Mutual Information-based Disentangled Neural Networks for Classifying Unseen Categories in Different Domains: Application to Fetal Ultrasound Imaging
Oct 30, 2020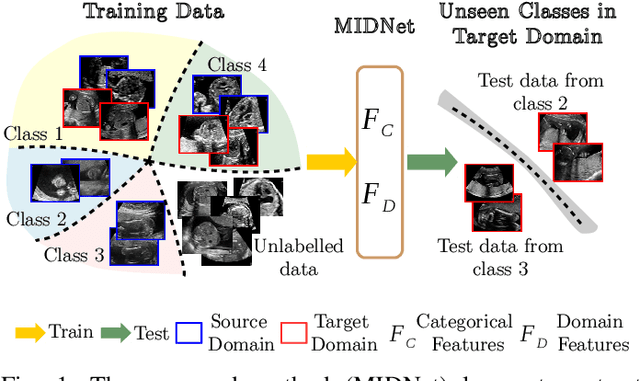
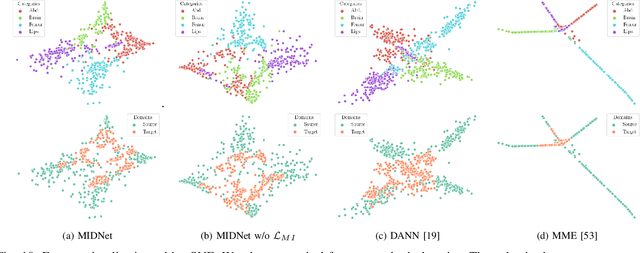
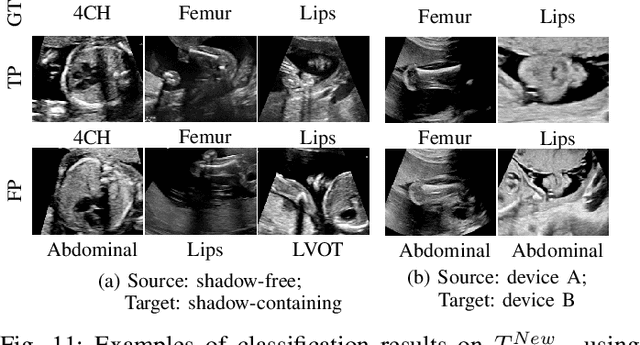
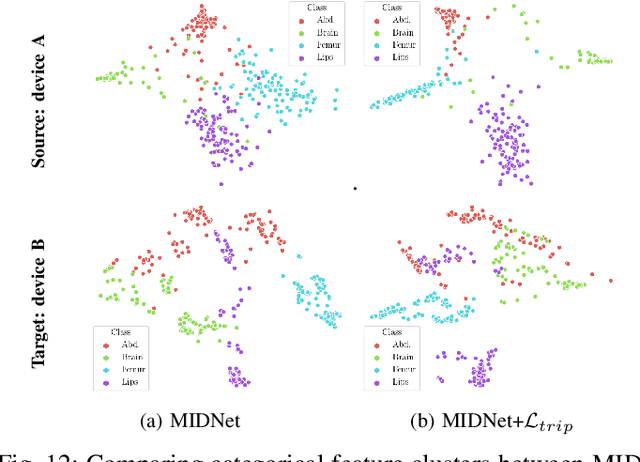
Abstract:Deep neural networks exhibit limited generalizability across images with different entangled domain features and categorical features. Learning generalizable features that can form universal categorical decision boundaries across domains is an interesting and difficult challenge. This problem occurs frequently in medical imaging applications when attempts are made to deploy and improve deep learning models across different image acquisition devices, across acquisition parameters or if some classes are unavailable in new training databases. To address this problem, we propose Mutual Information-based Disentangled Neural Networks (MIDNet), which extract generalizable categorical features to transfer knowledge to unseen categories in a target domain. The proposed MIDNet adopts a semi-supervised learning paradigm to alleviate the dependency on labeled data. This is important for real-world applications where data annotation is time-consuming, costly and requires training and expertise. We extensively evaluate the proposed method on fetal ultrasound datasets for two different image classification tasks where domain features are respectively defined by shadow artifacts and image acquisition devices. Experimental results show that the proposed method outperforms the state-of-the-art on the classification of unseen categories in a target domain with sparsely labeled training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge