Yunhe Gao
A Multi-scale Transformer for Medical Image Segmentation: Architectures, Model Efficiency, and Benchmarks
Mar 03, 2022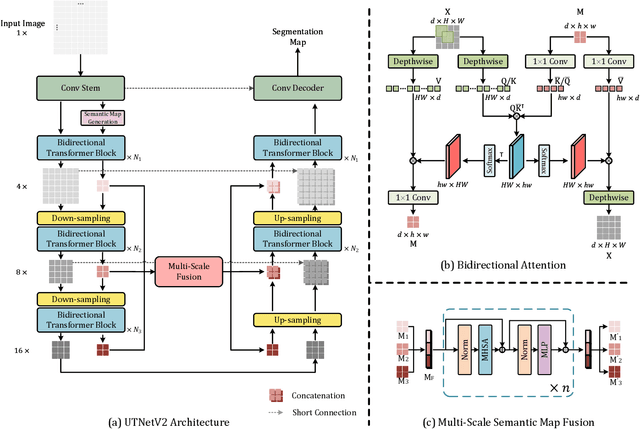
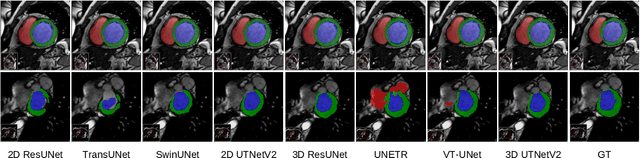
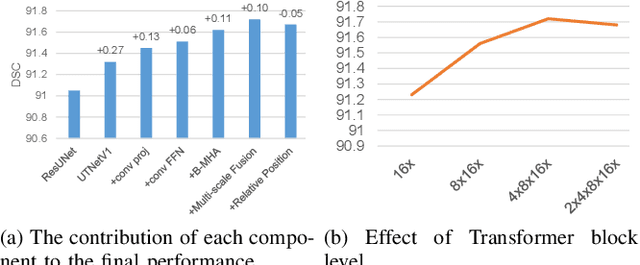
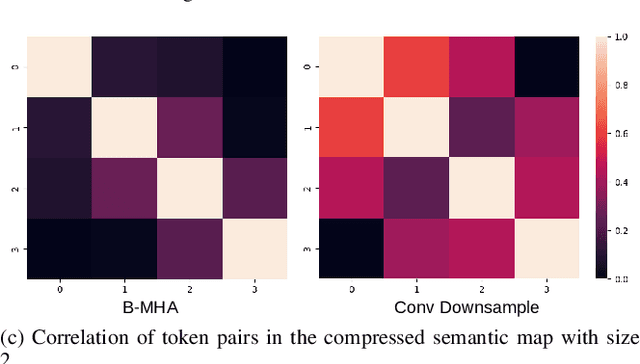
Abstract:Transformers have emerged to be successful in a number of natural language processing and vision tasks, but their potential applications to medical imaging remain largely unexplored due to the unique difficulties of this field. In this study, we present UTNetV2, a simple yet powerful backbone model that combines the strengths of the convolutional neural network and Transformer for enhancing performance and efficiency in medical image segmentation. The critical design of UTNetV2 includes three innovations: (1) We used a hybrid hierarchical architecture by introducing depthwise separable convolution to projection and feed-forward network in the Transformer block, which brings local relationship modeling and desirable properties of CNNs (translation invariance) to Transformer, thus eliminate the requirement of large-scale pre-training. (2) We proposed efficient bidirectional attention (B-MHA) that reduces the quadratic computation complexity of self-attention to linear by introducing an adaptively updated semantic map. The efficient attention makes it possible to capture long-range relationship and correct the fine-grained errors in high-resolution token maps. (3) The semantic maps in the B-MHA allow us to perform semantically and spatially global multi-scale feature fusion without introducing much computational overhead. Furthermore, we provide a fair comparison codebase of CNN-based and Transformer-based on various medical image segmentation tasks to evaluate the merits and defects of both architectures. UTNetV2 demonstrated state-of-the-art performance across various settings, including large-scale datasets, small-scale datasets, 2D and 3D settings.
Modality Bank: Learn multi-modality images across data centers without sharing medical data
Jan 22, 2022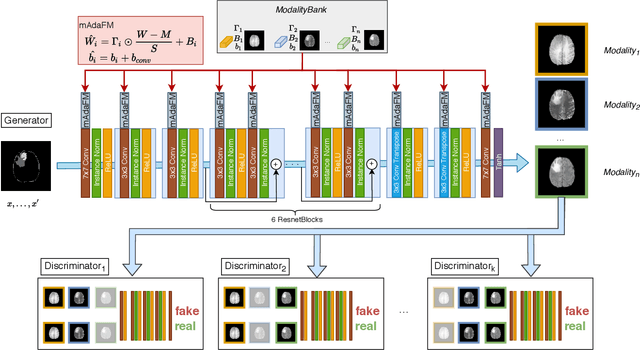
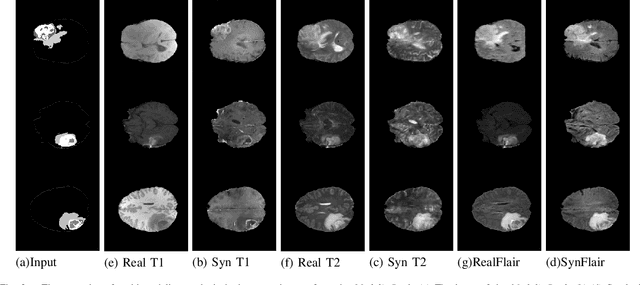
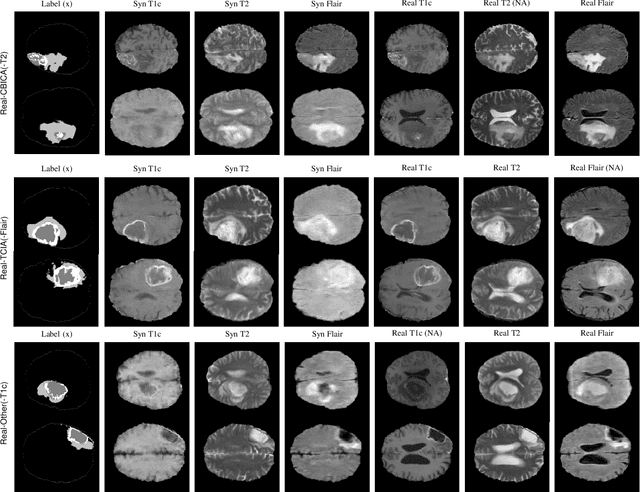
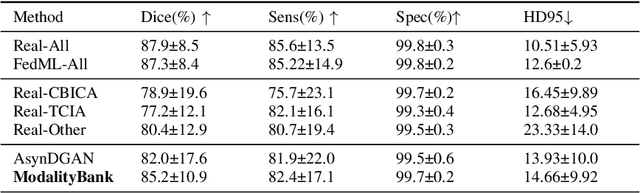
Abstract:Multi-modality images have been widely used and provide comprehensive information for medical image analysis. However, acquiring all modalities among all institutes is costly and often impossible in clinical settings. To leverage more comprehensive multi-modality information, we propose a privacy secured decentralized multi-modality adaptive learning architecture named ModalityBank. Our method could learn a set of effective domain-specific modulation parameters plugged into a common domain-agnostic network. We demonstrate by switching different sets of configurations, the generator could output high-quality images for a specific modality. Our method could also complete the missing modalities across all data centers, thus could be used for modality completion purposes. The downstream task trained from the synthesized multi-modality samples could achieve higher performance than learning from one real data center and achieve close-to-real performance compare with all real images.
UTNet: A Hybrid Transformer Architecture for Medical Image Segmentation
Jul 02, 2021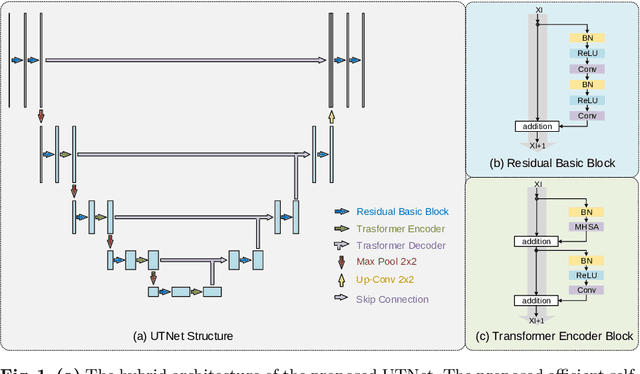
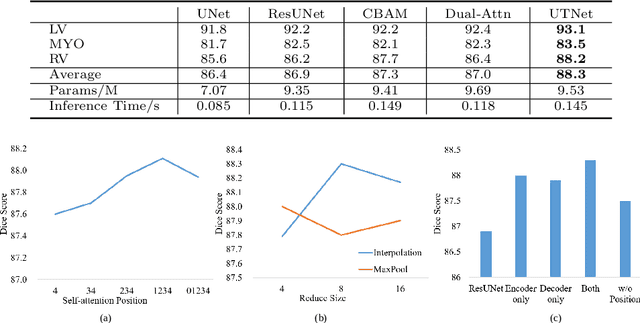
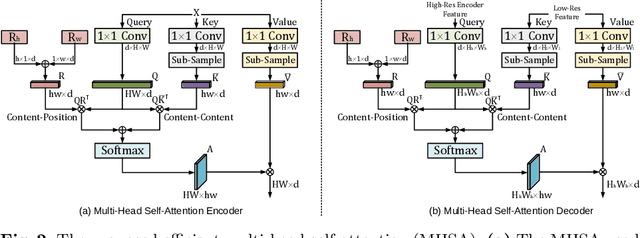
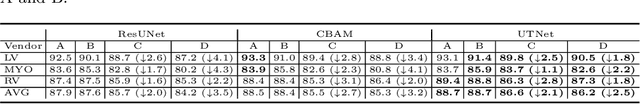
Abstract:Transformer architecture has emerged to be successful in a number of natural language processing tasks. However, its applications to medical vision remain largely unexplored. In this study, we present UTNet, a simple yet powerful hybrid Transformer architecture that integrates self-attention into a convolutional neural network for enhancing medical image segmentation. UTNet applies self-attention modules in both encoder and decoder for capturing long-range dependency at different scales with minimal overhead. To this end, we propose an efficient self-attention mechanism along with relative position encoding that reduces the complexity of self-attention operation significantly from $O(n^2)$ to approximate $O(n)$. A new self-attention decoder is also proposed to recover fine-grained details from the skipped connections in the encoder. Our approach addresses the dilemma that Transformer requires huge amounts of data to learn vision inductive bias. Our hybrid layer design allows the initialization of Transformer into convolutional networks without a need of pre-training. We have evaluated UTNet on the multi-label, multi-vendor cardiac magnetic resonance imaging cohort. UTNet demonstrates superior segmentation performance and robustness against the state-of-the-art approaches, holding the promise to generalize well on other medical image segmentations.
FocusNetv2: Imbalanced Large and Small Organ Segmentation with Adversarial Shape Constraint for Head and Neck CT Images
Apr 05, 2021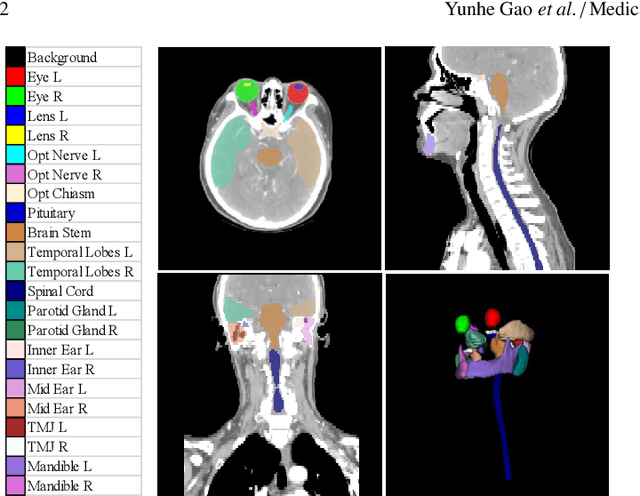
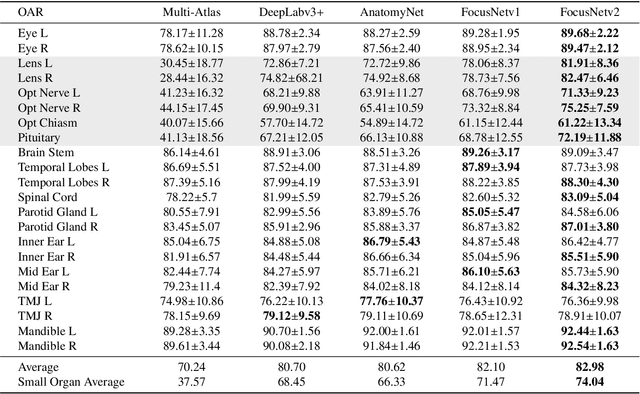
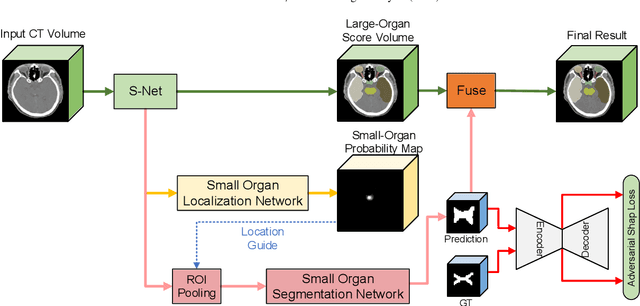
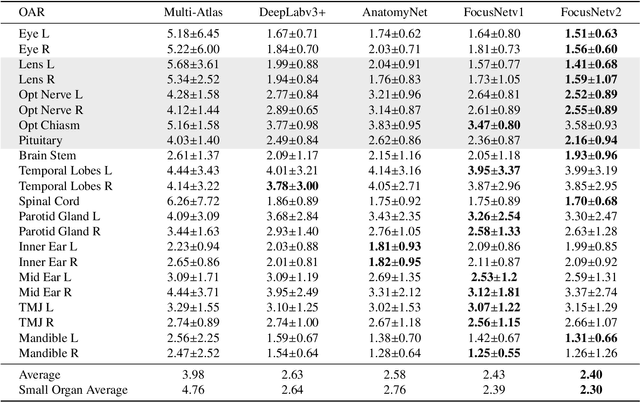
Abstract:Radiotherapy is a treatment where radiation is used to eliminate cancer cells. The delineation of organs-at-risk (OARs) is a vital step in radiotherapy treatment planning to avoid damage to healthy organs. For nasopharyngeal cancer, more than 20 OARs are needed to be precisely segmented in advance. The challenge of this task lies in complex anatomical structure, low-contrast organ contours, and the extremely imbalanced size between large and small organs. Common segmentation methods that treat them equally would generally lead to inaccurate small-organ labeling. We propose a novel two-stage deep neural network, FocusNetv2, to solve this challenging problem by automatically locating, ROI-pooling, and segmenting small organs with specifically designed small-organ localization and segmentation sub-networks while maintaining the accuracy of large organ segmentation. In addition to our original FocusNet, we employ a novel adversarial shape constraint on small organs to ensure the consistency between estimated small-organ shapes and organ shape prior knowledge. Our proposed framework is extensively tested on both self-collected dataset of 1,164 CT scans and the MICCAI Head and Neck Auto Segmentation Challenge 2015 dataset, which shows superior performance compared with state-of-the-art head and neck OAR segmentation methods.
Enabling Data Diversity: Efficient Automatic Augmentation via Regularized Adversarial Training
Mar 30, 2021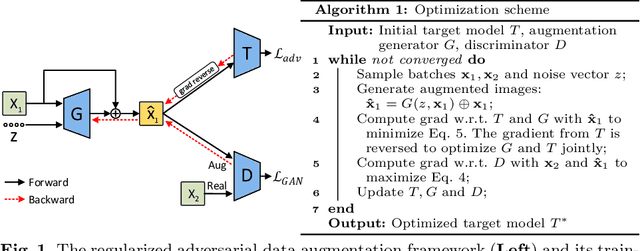

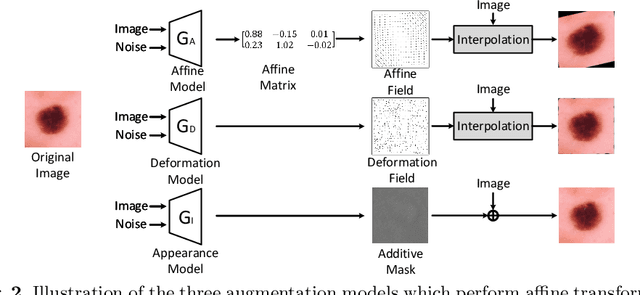
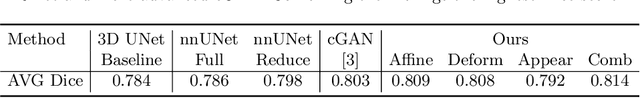
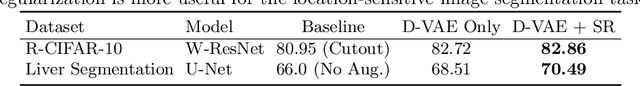
Abstract:Data augmentation has proved extremely useful by increasing training data variance to alleviate overfitting and improve deep neural networks' generalization performance. In medical image analysis, a well-designed augmentation policy usually requires much expert knowledge and is difficult to generalize to multiple tasks due to the vast discrepancies among pixel intensities, image appearances, and object shapes in different medical tasks. To automate medical data augmentation, we propose a regularized adversarial training framework via two min-max objectives and three differentiable augmentation models covering affine transformation, deformation, and appearance changes. Our method is more automatic and efficient than previous automatic augmentation methods, which still rely on pre-defined operations with human-specified ranges and costly bi-level optimization. Extensive experiments demonstrated that our approach, with less training overhead, achieves superior performance over state-of-the-art auto-augmentation methods on both tasks of 2D skin cancer classification and 3D organs-at-risk segmentation.
SelfNorm and CrossNorm for Out-of-Distribution Robustness
Feb 04, 2021
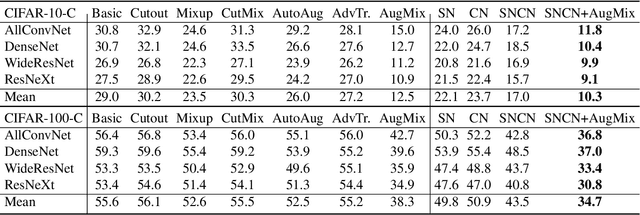
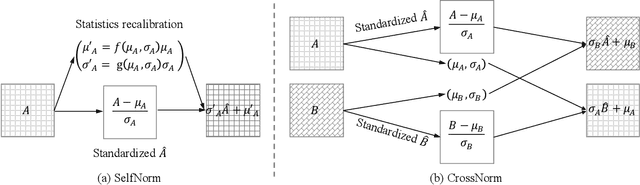
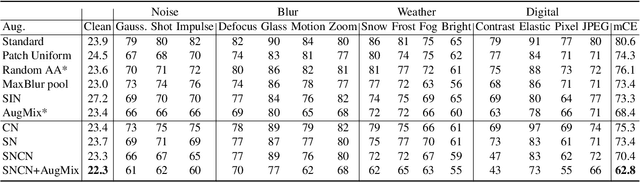
Abstract:Normalization techniques are crucial in stabilizing and accelerating the training of deep neural networks. However, they are mainly designed for the independent and identically distributed (IID) data, not satisfying many real-world out-of-distribution (OOD) situations. Unlike most previous works, this paper presents two normalization methods, SelfNorm and CrossNorm, to promote OOD generalization. SelfNorm uses attention to recalibrate statistics (channel-wise mean and variance), while CrossNorm exchanges the statistics between feature maps. SelfNorm and CrossNorm can complement each other in OOD generalization, though exploring different directions in statistics usage. Extensive experiments on different domains (vision and language), tasks (classification and segmentation), and settings (supervised and semi-supervised) show their effectiveness.
OnlineAugment: Online Data Augmentation with Less Domain Knowledge
Aug 22, 2020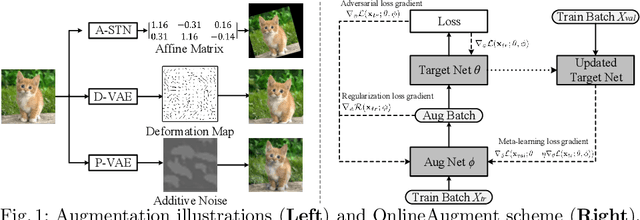
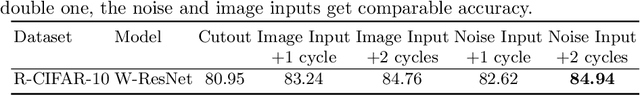
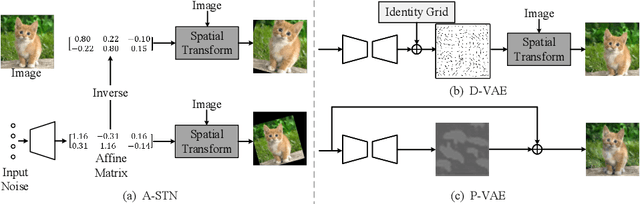
Abstract:Data augmentation is one of the most important tools in training modern deep neural networks. Recently, great advances have been made in searching for optimal augmentation policies in the image classification domain. However, two key points related to data augmentation remain uncovered by the current methods. First is that most if not all modern augmentation search methods are offline and learning policies are isolated from their usage. The learned policies are mostly constant throughout the training process and are not adapted to the current training model state. Second, the policies rely on class-preserving image processing functions. Hence applying current offline methods to new tasks may require domain knowledge to specify such kind of operations. In this work, we offer an orthogonal online data augmentation scheme together with three new augmentation networks, co-trained with the target learning task. It is both more efficient, in the sense that it does not require expensive offline training when entering a new domain, and more adaptive as it adapts to the learner state. Our augmentation networks require less domain knowledge and are easily applicable to new tasks. Extensive experiments demonstrate that the proposed scheme alone performs on par with the state-of-the-art offline data augmentation methods, as well as improving upon the state-of-the-art in combination with those methods. Code is available at https://github.com/zhiqiangdon/online-augment .
FocusNet: Imbalanced Large and Small Organ Segmentation with an End-to-End Deep Neural Network for Head and Neck CT Images
Jul 28, 2019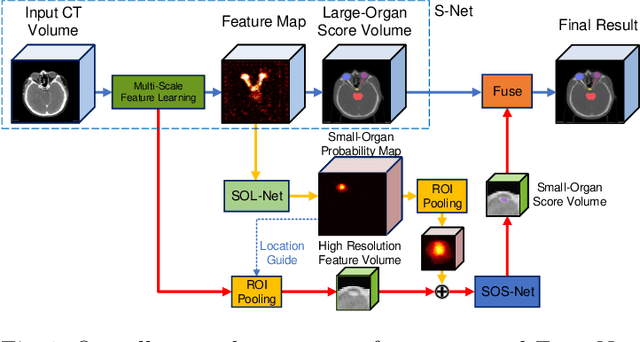
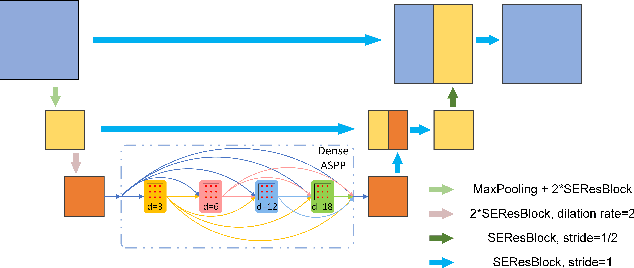
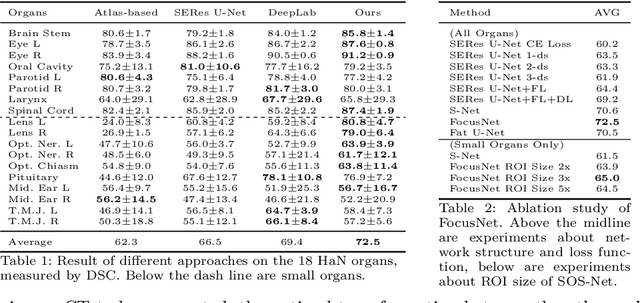
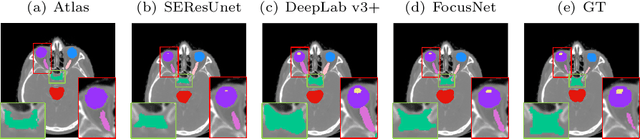
Abstract:In this paper, we propose an end-to-end deep neural network for solving the problem of imbalanced large and small organ segmentation in head and neck (HaN) CT images. To conduct radiotherapy planning for nasopharyngeal cancer, more than 10 organs-at-risk (normal organs) need to be precisely segmented in advance. However, the size ratio between large and small organs in the head could reach hundreds. Directly using such imbalanced organ annotations to train deep neural networks generally leads to inaccurate small-organ label maps. We propose a novel end-to-end deep neural network to solve this challenging problem by automatically locating, ROI-pooling, and segmenting small organs with specifically designed small-organ sub-networks while maintaining the accuracy of large organ segmentation. A strong main network with densely connected atrous spatial pyramid pooling and squeeze-and-excitation modules is used for segmenting large organs, where large organs' label maps are directly output. For small organs, their probabilistic locations instead of label maps are estimated by the main network. High-resolution and multi-scale feature volumes for each small organ are ROI-pooled according to their locations and are fed into small-organ networks for accurate segmenting small organs. Our proposed network is extensively tested on both collected real data and the \emph{MICCAI Head and Neck Auto Segmentation Challenge 2015} dataset, and shows superior performance compared with state-of-the-art segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge