Xiaowei Xu
How to Efficiently Adapt Large Segmentation Model(SAM) to Medical Images
Jun 23, 2023



Abstract:The emerging scale segmentation model, Segment Anything (SAM), exhibits impressive capabilities in zero-shot segmentation for natural images. However, when applied to medical images, SAM suffers from noticeable performance drop. To make SAM a real ``foundation model" for the computer vision community, it is critical to find an efficient way to customize SAM for medical image dataset. In this work, we propose to freeze SAM encoder and finetune a lightweight task-specific prediction head, as most of weights in SAM are contributed by the encoder. In addition, SAM is a promptable model, while prompt is not necessarily available in all application cases, and precise prompts for multiple class segmentation are also time-consuming. Therefore, we explore three types of prompt-free prediction heads in this work, include ViT, CNN, and linear layers. For ViT head, we remove the prompt tokens in the mask decoder of SAM, which is named AutoSAM. AutoSAM can also generate masks for different classes with one single inference after modification. To evaluate the label-efficiency of our finetuning method, we compare the results of these three prediction heads on a public medical image segmentation dataset with limited labeled data. Experiments demonstrate that finetuning SAM significantly improves its performance on medical image dataset, even with just one labeled volume. Moreover, AutoSAM and CNN prediction head also has better segmentation accuracy than training from scratch and self-supervised learning approaches when there is a shortage of annotations.
Additional Positive Enables Better Representation Learning for Medical Images
May 31, 2023


Abstract:This paper presents a new way to identify additional positive pairs for BYOL, a state-of-the-art (SOTA) self-supervised learning framework, to improve its representation learning ability. Unlike conventional BYOL which relies on only one positive pair generated by two augmented views of the same image, we argue that information from different images with the same label can bring more diversity and variations to the target features, thus benefiting representation learning. To identify such pairs without any label, we investigate TracIn, an instance-based and computationally efficient influence function, for BYOL training. Specifically, TracIn is a gradient-based method that reveals the impact of a training sample on a test sample in supervised learning. We extend it to the self-supervised learning setting and propose an efficient batch-wise per-sample gradient computation method to estimate the pairwise TracIn to represent the similarity of samples in the mini-batch during training. For each image, we select the most similar sample from other images as the additional positive and pull their features together with BYOL loss. Experimental results on two public medical datasets (i.e., ISIC 2019 and ChestX-ray) demonstrate that the proposed method can improve the classification performance compared to other competitive baselines in both semi-supervised and transfer learning settings.
TinyML Design Contest for Life-Threatening Ventricular Arrhythmia Detection
May 23, 2023Abstract:The first ACM/IEEE TinyML Design Contest (TDC) held at the 41st International Conference on Computer-Aided Design (ICCAD) in 2022 is a challenging, multi-month, research and development competition. TDC'22 focuses on real-world medical problems that require the innovation and implementation of artificial intelligence/machine learning (AI/ML) algorithms on implantable devices. The challenge problem of TDC'22 is to develop a novel AI/ML-based real-time detection algorithm for life-threatening ventricular arrhythmia over low-power microcontrollers utilized in Implantable Cardioverter-Defibrillators (ICDs). The dataset contains more than 38,000 5-second intracardiac electrograms (IEGMs) segments over 8 different types of rhythm from 90 subjects. The dedicated hardware platform is NUCLEO-L432KC manufactured by STMicroelectronics. TDC'22, which is open to multi-person teams world-wide, attracted more than 150 teams from over 50 organizations. This paper first presents the medical problem, dataset, and evaluation procedure in detail. It further demonstrates and discusses the designs developed by the leading teams as well as representative results. This paper concludes with the direction of improvement for the future TinyML design for health monitoring applications.
SATBA: An Invisible Backdoor Attack Based On Spatial Attention
Feb 25, 2023Abstract:As a new realm of AI security, backdoor attack has drew growing attention research in recent years. It is well known that backdoor can be injected in a DNN model through the process of model training with poisoned dataset which is consist of poisoned sample. The injected model output correct prediction on benign samples yet behave abnormally on poisoned samples included trigger pattern. Most existing trigger of poisoned sample are visible and can be easily found by human visual inspection, and the trigger injection process will cause the feature loss of natural sample and trigger. To solve the above problems and inspire by spatial attention mechanism, we introduce a novel backdoor attack named SATBA, which is invisible and can minimize the loss of trigger to improve attack success rate and model accuracy. It extracts data features and generate trigger pattern related to clean data through spatial attention, poisons clean image by using a U-type models to plant a trigger into the original data. We demonstrate the effectiveness of our attack against three popular image classification DNNs on three standard datasets. Besides, we conduct extensive experiments about image similarity to show that our proposed attack can provide practical stealthiness which is critical to resist to backdoor defense.
ImageCAS: A Large-Scale Dataset and Benchmark for Coronary Artery Segmentation based on Computed Tomography Angiography Images
Nov 03, 2022Abstract:Cardiovascular disease (CVD) accounts for about half of non-communicable diseases. Vessel stenosis in the coronary artery is considered to be the major risk of CVD. Computed tomography angiography (CTA) is one of the widely used noninvasive imaging modalities in coronary artery diagnosis due to its superior image resolution. Clinically, segmentation of coronary arteries is essential for the diagnosis and quantification of coronary artery disease. Recently, a variety of works have been proposed to address this problem. However, on one hand, most works rely on in-house datasets, and only a few works published their datasets to the public which only contain tens of images. On the other hand, their source code have not been published, and most follow-up works have not made comparison with existing works, which makes it difficult to judge the effectiveness of the methods and hinders the further exploration of this challenging yet critical problem in the community. In this paper, we propose a large-scale dataset for coronary artery segmentation on CTA images. In addition, we have implemented a benchmark in which we have tried our best to implement several typical existing methods. Furthermore, we propose a strong baseline method which combines multi-scale patch fusion and two-stage processing to extract the details of vessels. Comprehensive experiments show that the proposed method achieves better performance than existing works on the proposed large-scale dataset. The benchmark and the dataset are published at https://github.com/XiaoweiXu/ImageCAS-A-Large-Scale-Dataset-and-Benchmark-for-Coronary-Artery-Segmentation-based-on-CT.
Unsupervised Knowledge Graph Construction and Event-centric Knowledge Infusion for Scientific NLI
Oct 28, 2022Abstract:With the advance of natural language inference (NLI), a rising demand for NLI is to handle scientific texts. Existing methods depend on pre-trained models (PTM) which lack domain-specific knowledge. To tackle this drawback, we introduce a scientific knowledge graph to generalize PTM to scientific domain. However, existing knowledge graph construction approaches suffer from some drawbacks, i.e., expensive labeled data, failure to apply in other domains, long inference time and difficulty extending to large corpora. Therefore, we propose an unsupervised knowledge graph construction method to build a scientific knowledge graph (SKG) without any labeled data. Moreover, to alleviate noise effect from SKG and complement knowledge in sentences better, we propose an event-centric knowledge infusion method to integrate external knowledge into each event that is a fine-grained semantic unit in sentences. Experimental results show that our method achieves state-of-the-art performance and the effectiveness and reliability of SKG.
Sub-cluster-aware Network for Few-shot Skin Disease Classification
Jul 03, 2022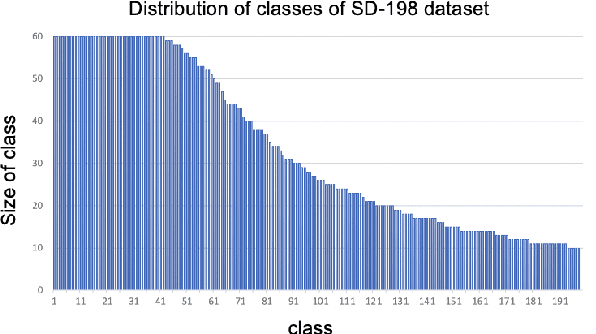
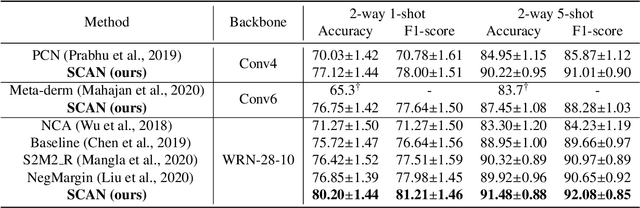
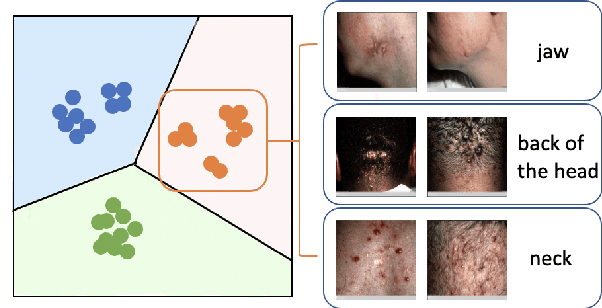
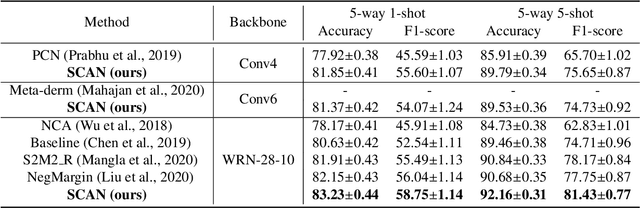
Abstract:This paper studies the few-shot skin disease classification problem. Based on a crucial observation that skin disease images often exist multiple sub-clusters within a class (i.e., the appearances of images within one class of disease vary and form multiple distinct sub-groups), we design a novel Sub-Cluster-Aware Network, namely SCAN, for rare skin disease diagnosis with enhanced accuracy. As the performance of few-shot learning highly depends on the quality of the learned feature encoder, the main principle guiding the design of SCAN is the intrinsic sub-clustered representation learning for each class so as to better describe feature distributions. Specifically, SCAN follows a dual-branch framework, where the first branch is to learn class-wise features to distinguish different skin diseases, and the second one aims to learn features which can effectively partition each class into several groups so as to preserve the sub-clustered structure within each class. To achieve the objective of the second branch, we present a cluster loss to learn image similarities via unsupervised clustering. To ensure that the samples in each sub-cluster are from the same class, we further design a purity loss to refine the unsupervised clustering results. We evaluate the proposed approach on two public datasets for few-shot skin disease classification. The experimental results validate that our framework outperforms the other state-of-the-art methods by around 2% to 4% on the SD-198 and Derm7pt datasets.
RT-DNAS: Real-time Constrained Differentiable Neural Architecture Search for 3D Cardiac Cine MRI Segmentation
Jun 13, 2022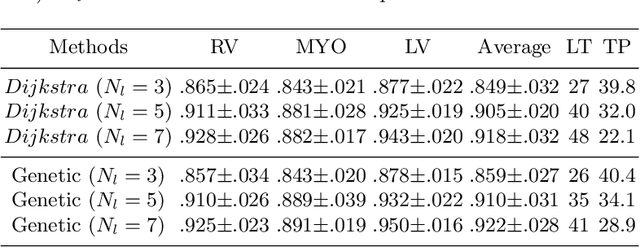

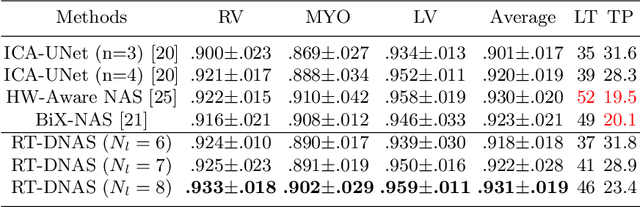
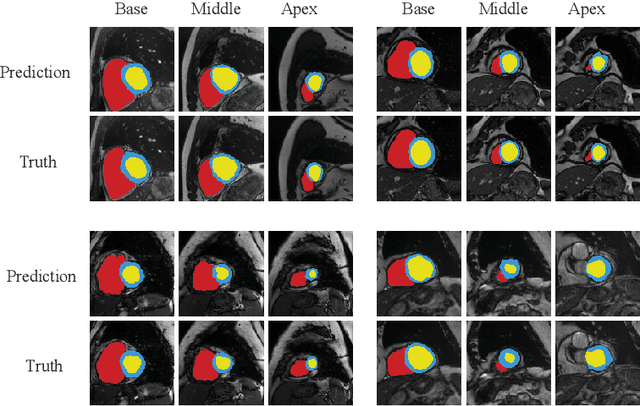
Abstract:Accurately segmenting temporal frames of cine magnetic resonance imaging (MRI) is a crucial step in various real-time MRI guided cardiac interventions. To achieve fast and accurate visual assistance, there are strict requirements on the maximum latency and minimum throughput of the segmentation framework. State-of-the-art neural networks on this task are mostly hand-crafted to satisfy these constraints while achieving high accuracy. On the other hand, while existing literature have demonstrated the power of neural architecture search (NAS) in automatically identifying the best neural architectures for various medical applications, they are mostly guided by accuracy, sometimes with computation complexity, and the importance of real-time constraints are overlooked. A major challenge is that such constraints are non-differentiable and are thus not compatible with the widely used differentiable NAS frameworks. In this paper, we present a strategy that directly handles real-time constraints in a differentiable NAS framework named RT-DNAS. Experiments on extended 2017 MICCAI ACDC dataset show that compared with state-of-the-art manually and automatically designed architectures, RT-DNAS is able to identify ones with better accuracy while satisfying the real-time constraints.
FairPrune: Achieving Fairness Through Pruning for Dermatological Disease Diagnosis
Mar 04, 2022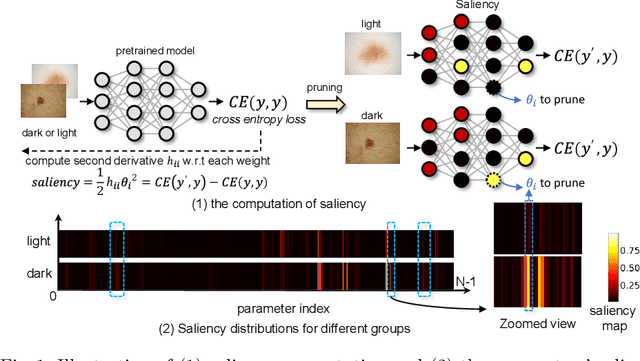
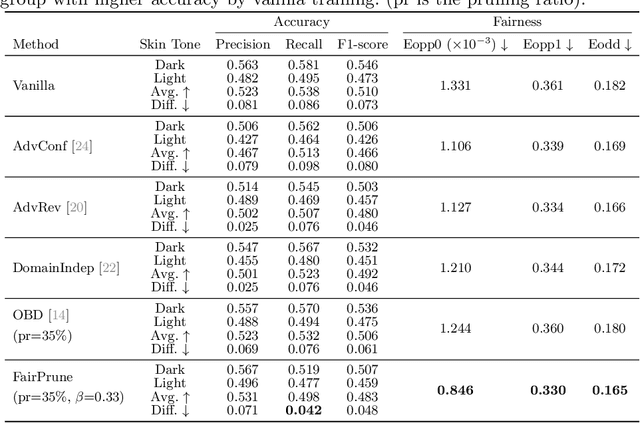
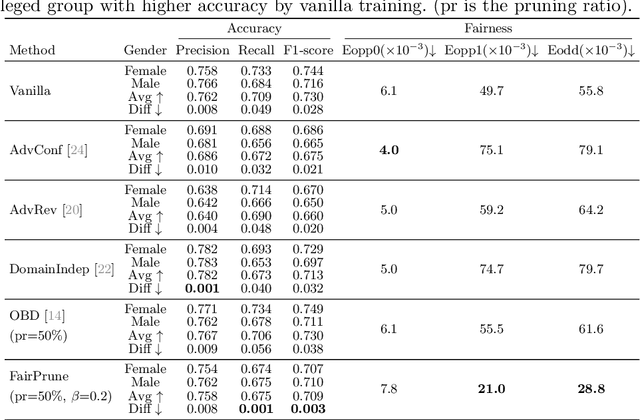
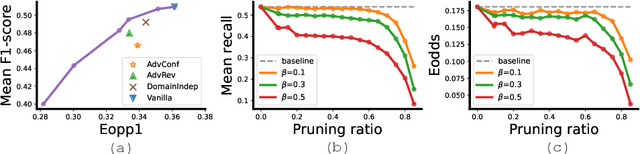
Abstract:Many works have shown that deep learning-based medical image classification models can exhibit bias toward certain demographic attributes like race, gender, and age. Existing bias mitigation methods primarily focus on learning debiased models, which may not necessarily guarantee all sensitive information can be removed and usually comes with considerable accuracy degradation on both privileged and unprivileged groups. To tackle this issue, we propose a method, FairPrune, that achieves fairness by pruning. Conventionally, pruning is used to reduce the model size for efficient inference. However, we show that pruning can also be a powerful tool to achieve fairness. Our observation is that during pruning, each parameter in the model has different importance for different groups' accuracy. By pruning the parameters based on this importance difference, we can reduce the accuracy difference between the privileged group and the unprivileged group to improve fairness without a large accuracy drop. To this end, we use the second derivative of the parameters of a pre-trained model to quantify the importance of each parameter with respect to the model accuracy for each group. Experiments on two skin lesion diagnosis datasets over multiple sensitive attributes demonstrate that our method can greatly improve fairness while keeping the average accuracy of both groups as high as possible.
"One-Shot" Reduction of Additive Artifacts in Medical Images
Oct 23, 2021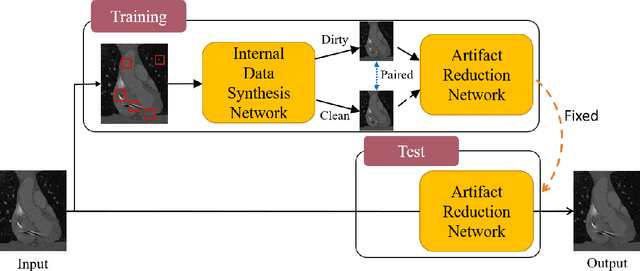
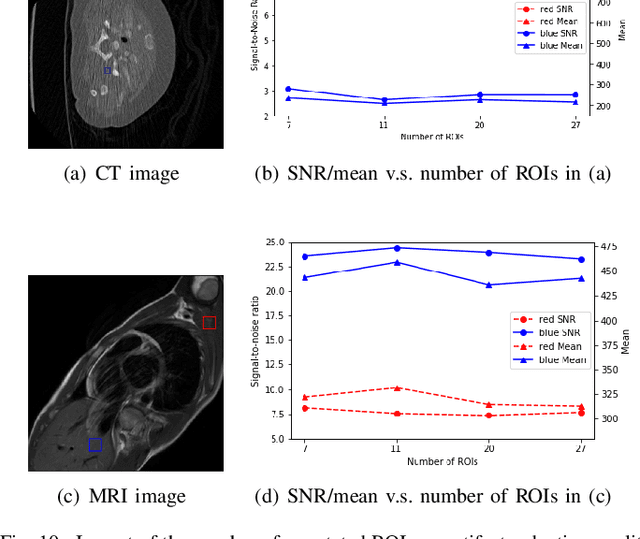
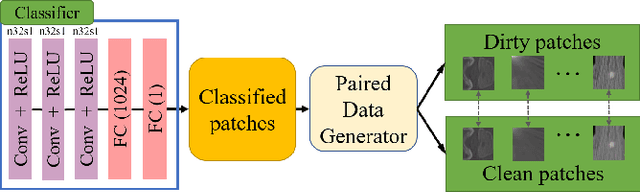
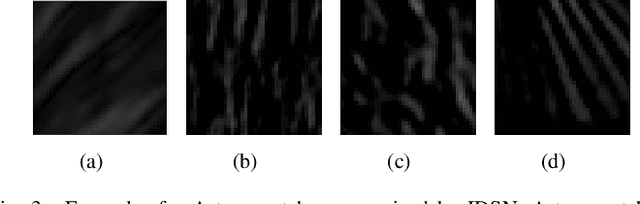
Abstract:Medical images may contain various types of artifacts with different patterns and mixtures, which depend on many factors such as scan setting, machine condition, patients' characteristics, surrounding environment, etc. However, existing deep-learning-based artifact reduction methods are restricted by their training set with specific predetermined artifact types and patterns. As such, they have limited clinical adoption. In this paper, we introduce One-Shot medical image Artifact Reduction (OSAR), which exploits the power of deep learning but without using pre-trained general networks. Specifically, we train a light-weight image-specific artifact reduction network using data synthesized from the input image at test-time. Without requiring any prior large training data set, OSAR can work with almost any medical images that contain varying additive artifacts which are not in any existing data sets. In addition, Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) are used as vehicles and show that the proposed method can reduce artifacts better than state-of-the-art both qualitatively and quantitatively using shorter test time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge