Xiaosong Wang
Going to Extremes: Weakly Supervised Medical Image Segmentation
Sep 25, 2020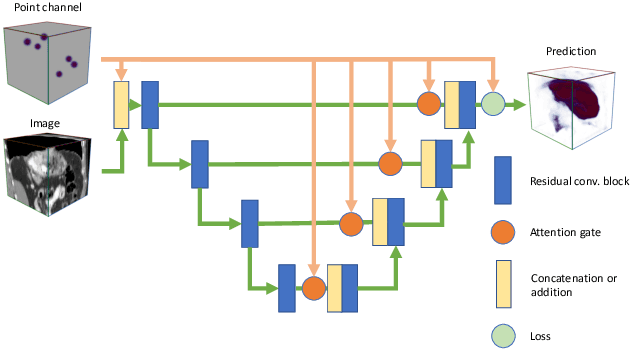
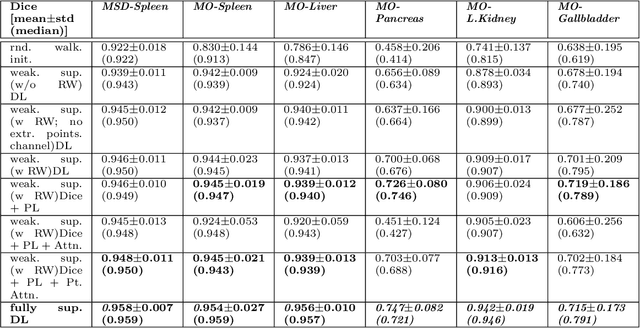
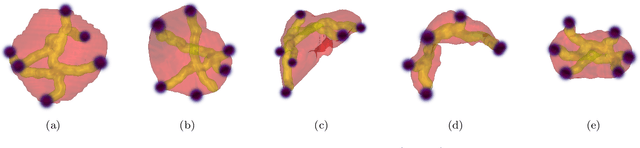
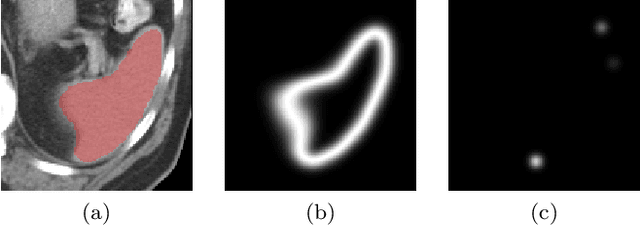
Abstract:Medical image annotation is a major hurdle for developing precise and robust machine learning models. Annotation is expensive, time-consuming, and often requires expert knowledge, particularly in the medical field. Here, we suggest using minimal user interaction in the form of extreme point clicks to train a segmentation model which, in effect, can be used to speed up medical image annotation. An initial segmentation is generated based on the extreme points utilizing the random walker algorithm. This initial segmentation is then used as a noisy supervision signal to train a fully convolutional network that can segment the organ of interest, based on the provided user clicks. Through experimentation on several medical imaging datasets, we show that the predictions of the network can be refined using several rounds of training with the prediction from the same weakly annotated data. Further improvements are shown utilizing the clicked points within a custom-designed loss and attention mechanism. Our approach has the potential to speed up the process of generating new training datasets for the development of new machine learning and deep learning-based models for, but not exclusively, medical image analysis.
Weakly supervised one-stage vision and language disease detection using large scale pneumonia and pneumothorax studies
Jul 31, 2020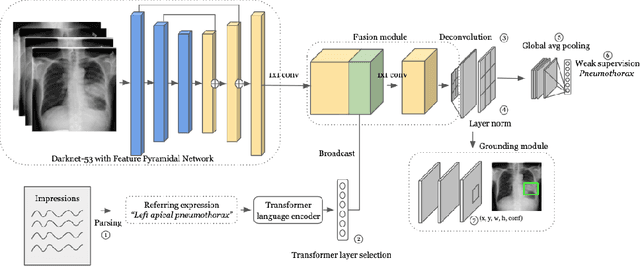
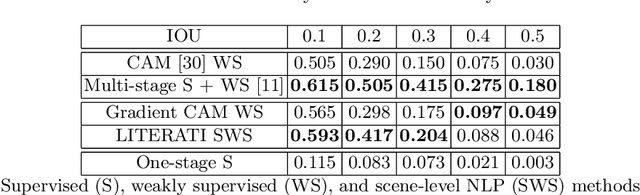
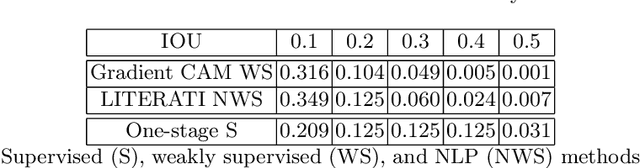
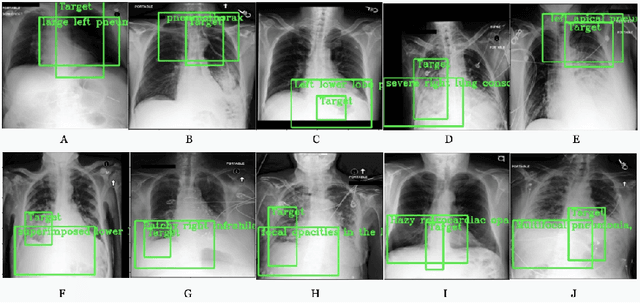
Abstract:Detecting clinically relevant objects in medical images is a challenge despite large datasets due to the lack of detailed labels. To address the label issue, we utilize the scene-level labels with a detection architecture that incorporates natural language information. We present a challenging new set of radiologist paired bounding box and natural language annotations on the publicly available MIMIC-CXR dataset especially focussed on pneumonia and pneumothorax. Along with the dataset, we present a joint vision language weakly supervised transformer layer-selected one-stage dual head detection architecture (LITERATI) alongside strong baseline comparisons with class activation mapping (CAM), gradient CAM, and relevant implementations on the NIH ChestXray-14 and MIMIC-CXR dataset. Borrowing from advances in vision language architectures, the LITERATI method demonstrates joint image and referring expression (objects localized in the image using natural language) input for detection that scales in a purely weakly supervised fashion. The architectural modifications address three obstacles -- implementing a supervised vision and language detection method in a weakly supervised fashion, incorporating clinical referring expression natural language information, and generating high fidelity detections with map probabilities. Nevertheless, the challenging clinical nature of the radiologist annotations including subtle references, multi-instance specifications, and relatively verbose underlying medical reports, ensures the vision language detection task at scale remains stimulating for future investigation.
Multi-Domain Image Completion for Random Missing Input Data
Jul 10, 2020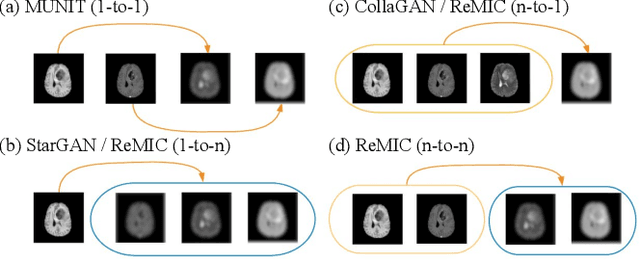
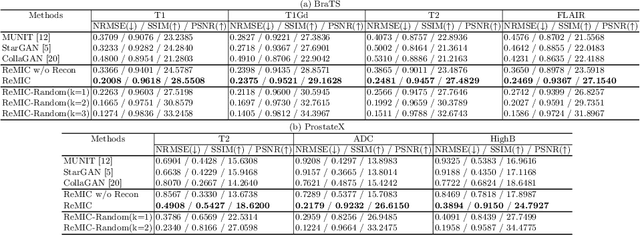
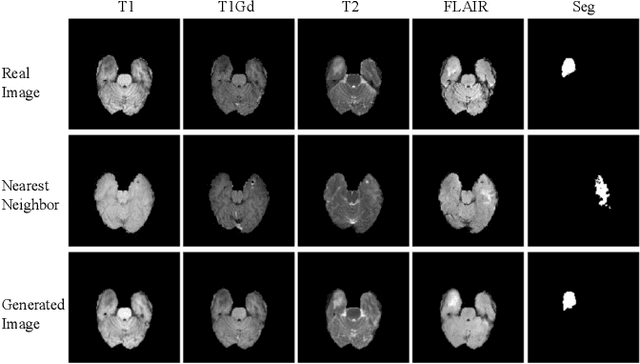
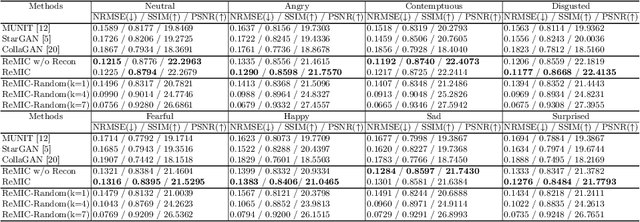
Abstract:Multi-domain data are widely leveraged in vision applications taking advantage of complementary information from different modalities, e.g., brain tumor segmentation from multi-parametric magnetic resonance imaging (MRI). However, due to possible data corruption and different imaging protocols, the availability of images for each domain could vary amongst multiple data sources in practice, which makes it challenging to build a universal model with a varied set of input data. To tackle this problem, we propose a general approach to complete the random missing domain(s) data in real applications. Specifically, we develop a novel multi-domain image completion method that utilizes a generative adversarial network (GAN) with a representational disentanglement scheme to extract shared skeleton encoding and separate flesh encoding across multiple domains. We further illustrate that the learned representation in multi-domain image completion could be leveraged for high-level tasks, e.g., segmentation, by introducing a unified framework consisting of image completion and segmentation with a shared content encoder. The experiments demonstrate consistent performance improvement on three datasets for brain tumor segmentation, prostate segmentation, and facial expression image completion respectively.
Enhancing Foreground Boundaries for Medical Image Segmentation
May 29, 2020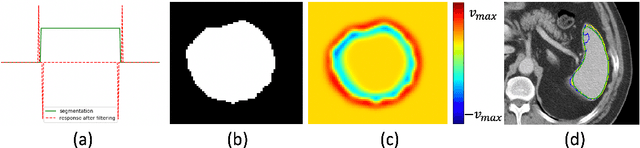
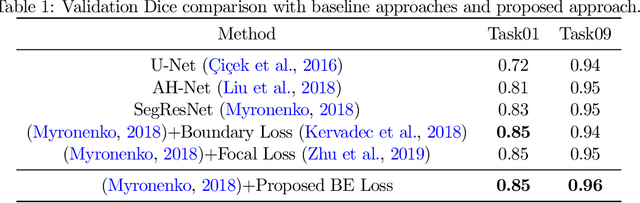
Abstract:Object segmentation plays an important role in the modern medical image analysis, which benefits clinical study, disease diagnosis, and surgery planning. Given the various modalities of medical images, the automated or semi-automated segmentation approaches have been used to identify and parse organs, bones, tumors, and other regions-of-interest (ROI). However, these contemporary segmentation approaches tend to fail to predict the boundary areas of ROI, because of the fuzzy appearance contrast caused during the imaging procedure. To further improve the segmentation quality of boundary areas, we propose a boundary enhancement loss to enforce additional constraints on optimizing machine learning models. The proposed loss function is light-weighted and easy to implement without any pre- or post-processing. Our experimental results validate that our loss function are better than, or at least comparable to, other state-of-the-art loss functions in terms of segmentation accuracy.
When Radiology Report Generation Meets Knowledge Graph
Feb 19, 2020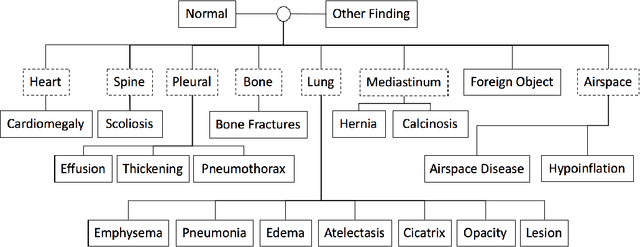
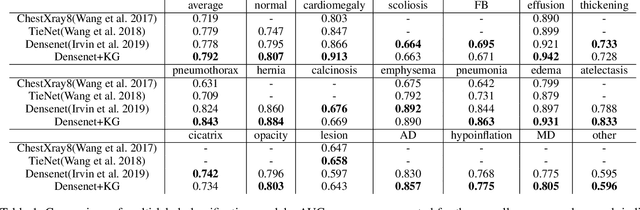
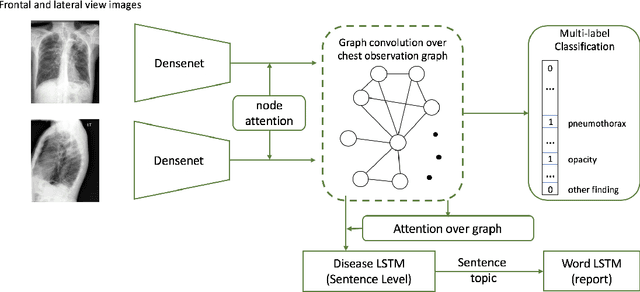
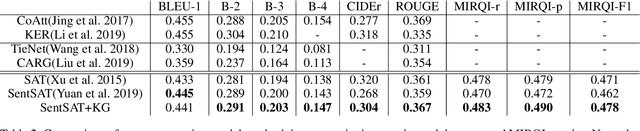
Abstract:Automatic radiology report generation has been an attracting research problem towards computer-aided diagnosis to alleviate the workload of doctors in recent years. Deep learning techniques for natural image captioning are successfully adapted to generating radiology reports. However, radiology image reporting is different from the natural image captioning task in two aspects: 1) the accuracy of positive disease keyword mentions is critical in radiology image reporting in comparison to the equivalent importance of every single word in a natural image caption; 2) the evaluation of reporting quality should focus more on matching the disease keywords and their associated attributes instead of counting the occurrence of N-gram. Based on these concerns, we propose to utilize a pre-constructed graph embedding module (modeled with a graph convolutional neural network) on multiple disease findings to assist the generation of reports in this work. The incorporation of knowledge graph allows for dedicated feature learning for each disease finding and the relationship modeling between them. In addition, we proposed a new evaluation metric for radiology image reporting with the assistance of the same composed graph. Experimental results demonstrate the superior performance of the methods integrated with the proposed graph embedding module on a publicly accessible dataset (IU-RR) of chest radiographs compared with previous approaches using both the conventional evaluation metrics commonly adopted for image captioning and our proposed ones.
Weakly supervised segmentation from extreme points
Oct 02, 2019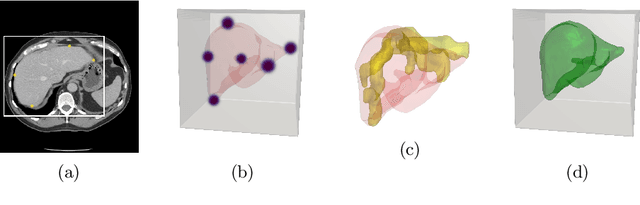
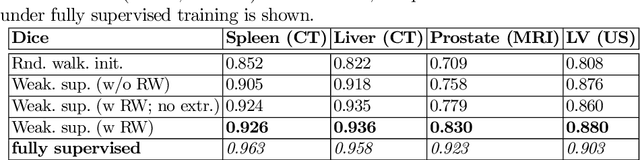
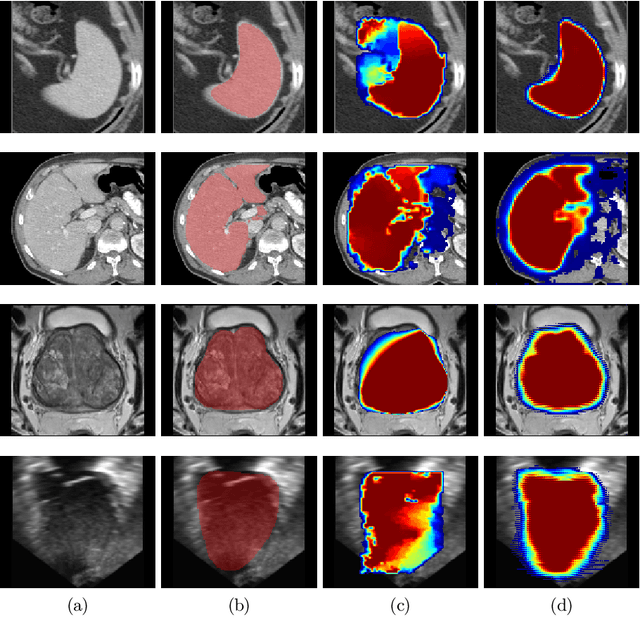
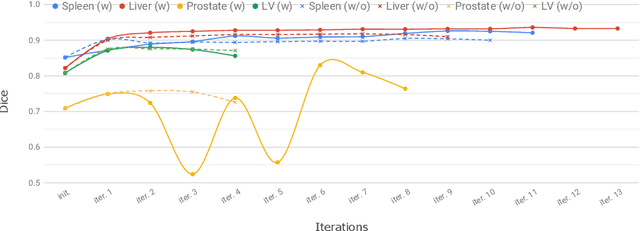
Abstract:Annotation of medical images has been a major bottleneck for the development of accurate and robust machine learning models. Annotation is costly and time-consuming and typically requires expert knowledge, especially in the medical domain. Here, we propose to use minimal user interaction in the form of extreme point clicks in order to train a segmentation model that can, in turn, be used to speed up the annotation of medical images. We use extreme points in each dimension of a 3D medical image to constrain an initial segmentation based on the random walker algorithm. This segmentation is then used as a weak supervisory signal to train a fully convolutional network that can segment the organ of interest based on the provided user clicks. We show that the network's predictions can be refined through several iterations of training and prediction using the same weakly annotated data. Ultimately, our method has the potential to speed up the generation process of new training datasets for the development of new machine learning and deep learning-based models for, but not exclusively, medical image analysis.
Correlation via synthesis: end-to-end nodule image generation and radiogenomic map learning based on generative adversarial network
Jul 08, 2019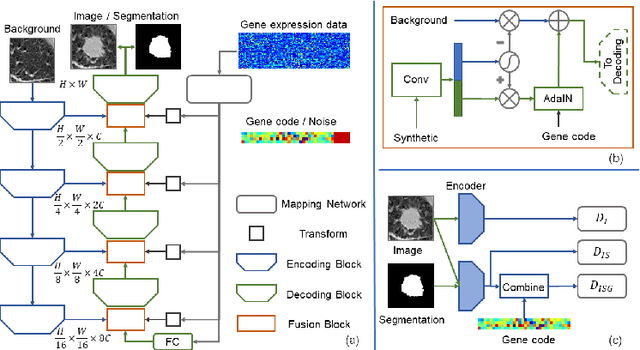

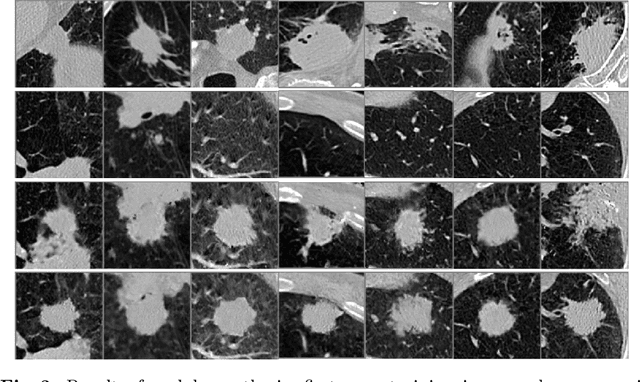
Abstract:Radiogenomic map linking image features and gene expression profiles is useful for noninvasively identifying molecular properties of a particular type of disease. Conventionally, such map is produced in three separate steps: 1) gene-clustering to "metagenes", 2) image feature extraction, and 3) statistical correlation between metagenes and image features. Each step is independently performed and relies on arbitrary measurements. In this work, we investigate the potential of an end-to-end method fusing gene data with image features to generate synthetic image and learn radiogenomic map simultaneously. To achieve this goal, we develop a generative adversarial network (GAN) conditioned on both background images and gene expression profiles, synthesizing the corresponding image. Image and gene features are fused at different scales to ensure the realism and quality of the synthesized image. We tested our method on non-small cell lung cancer (NSCLC) dataset. Results demonstrate that the proposed method produces realistic synthetic images, and provides a promising way to find gene-image relationship in a holistic end-to-end manner.
When Unseen Domain Generalization is Unnecessary? Rethinking Data Augmentation
Jun 12, 2019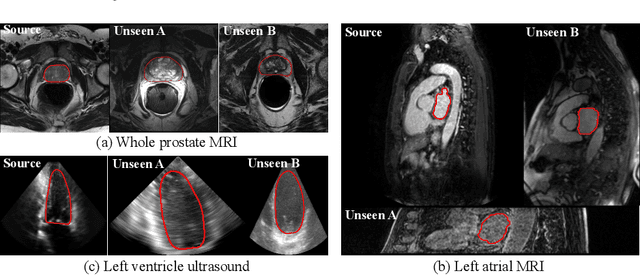
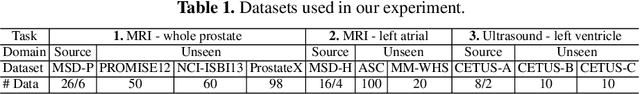
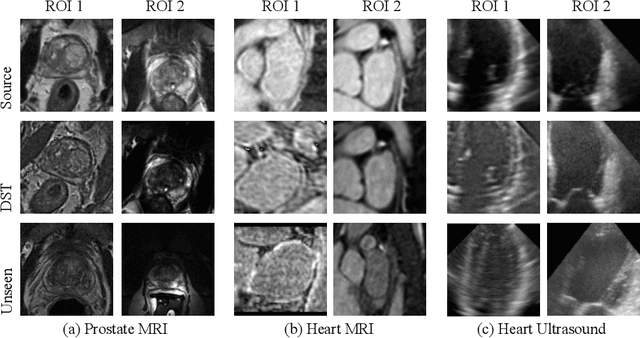
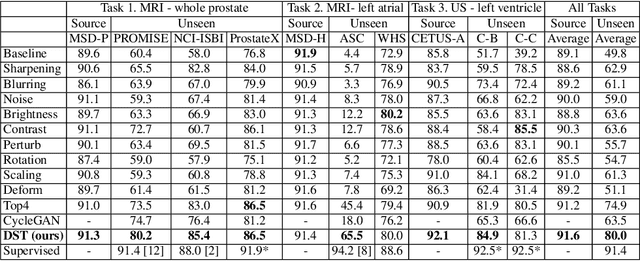
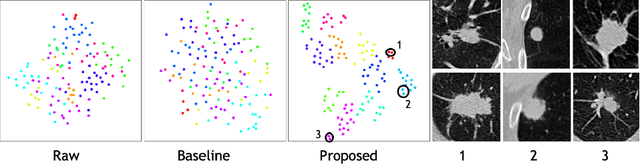
Abstract:Recent advances in deep learning for medical image segmentation demonstrate expert-level accuracy. However, in clinically realistic environments, such methods have marginal performance due to differences in image domains, including different imaging protocols, device vendors and patient populations. Here we consider the problem of domain generalization, when a model is trained once, and its performance generalizes to unseen domains. Intuitively, within a specific medical imaging modality the domain differences are smaller relative to natural images domain variability. We rethink data augmentation for medical 3D images and propose a deep stacked transformations (DST) approach for domain generalization. Specifically, a series of n stacked transformations are applied to each image in each mini-batch during network training to account for the contribution of domain-specific shifts in medical images. We comprehensively evaluate our method on three tasks: segmentation of whole prostate from 3D MRI, left atrial from 3D MRI, and left ventricle from 3D ultrasound. We demonstrate that when trained on a small source dataset, (i) on average, DST models on unseen datasets degrade only by 11% (Dice score change), compared to the conventional augmentation (degrading 39%) and CycleGAN-based domain adaptation method (degrading 25%); (ii) when evaluation on the same domain, DST is also better albeit only marginally. (iii) When training on large-sized data, DST on unseen domains reaches performance of state-of-the-art fully supervised models. These findings establish a strong benchmark for the study of domain generalization in medical imaging, and can be generalized to the design of robust deep segmentation models for clinical deployment.
Deep Lesion Graphs in the Wild: Relationship Learning and Organization of Significant Radiology Image Findings in a Diverse Large-scale Lesion Database
Jul 28, 2018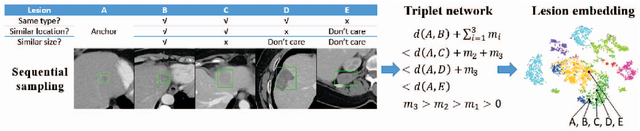
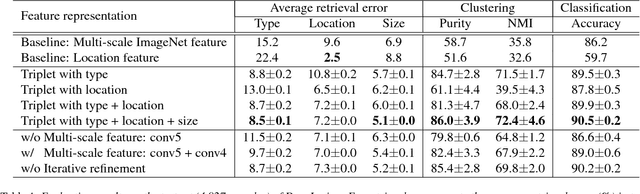
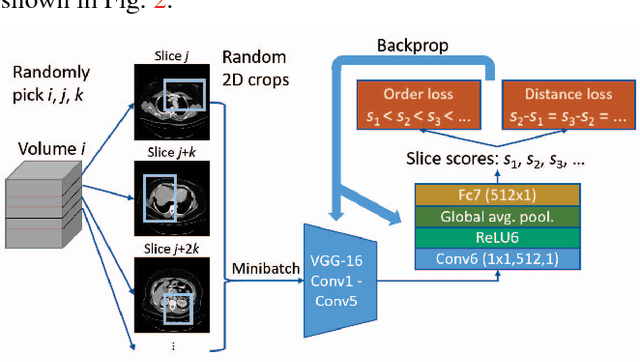
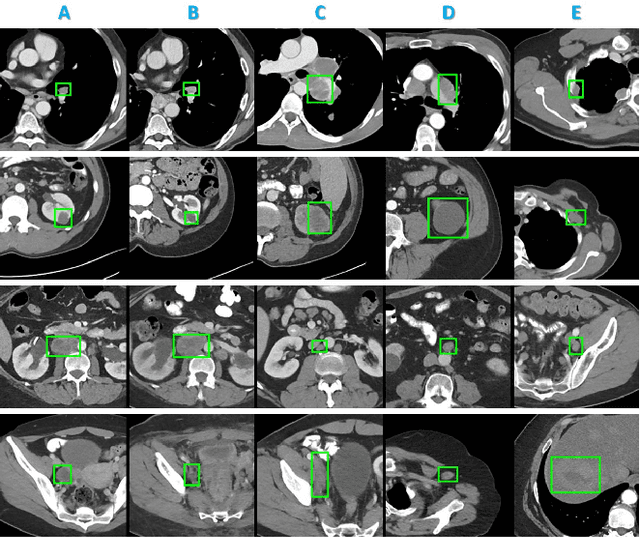
Abstract:Radiologists in their daily work routinely find and annotate significant abnormalities on a large number of radiology images. Such abnormalities, or lesions, have collected over years and stored in hospitals' picture archiving and communication systems. However, they are basically unsorted and lack semantic annotations like type and location. In this paper, we aim to organize and explore them by learning a deep feature representation for each lesion. A large-scale and comprehensive dataset, DeepLesion, is introduced for this task. DeepLesion contains bounding boxes and size measurements of over 32K lesions. To model their similarity relationship, we leverage multiple supervision information including types, self-supervised location coordinates and sizes. They require little manual annotation effort but describe useful attributes of the lesions. Then, a triplet network is utilized to learn lesion embeddings with a sequential sampling strategy to depict their hierarchical similarity structure. Experiments show promising qualitative and quantitative results on lesion retrieval, clustering, and classification. The learned embeddings can be further employed to build a lesion graph for various clinically useful applications. We propose algorithms for intra-patient lesion matching and missing annotation mining. Experimental results validate their effectiveness.
Attention-Guided Curriculum Learning for Weakly Supervised Classification and Localization of Thoracic Diseases on Chest Radiographs
Jul 19, 2018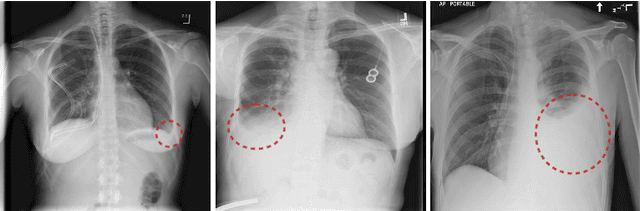
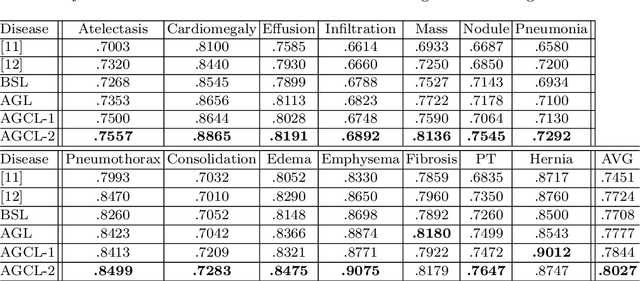
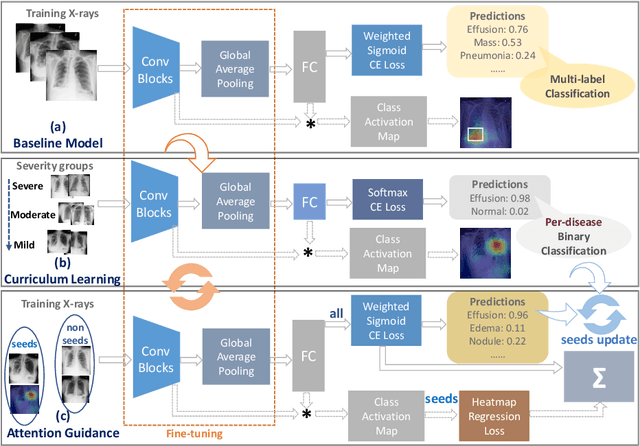
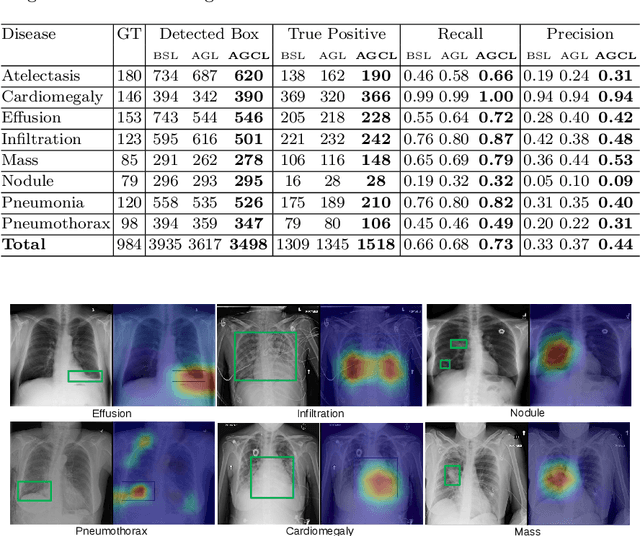
Abstract:In this work, we exploit the task of joint classification and weakly supervised localization of thoracic diseases from chest radiographs, with only image-level disease labels coupled with disease severity-level (DSL) information of a subset. A convolutional neural network (CNN) based attention-guided curriculum learning (AGCL) framework is presented, which leverages the severity-level attributes mined from radiology reports. Images in order of difficulty (grouped by different severity-levels) are fed to CNN to boost the learning gradually. In addition, highly confident samples (measured by classification probabilities) and their corresponding class-conditional heatmaps (generated by the CNN) are extracted and further fed into the AGCL framework to guide the learning of more distinctive convolutional features in the next iteration. A two-path network architecture is designed to regress the heatmaps from selected seed samples in addition to the original classification task. The joint learning scheme can improve the classification and localization performance along with more seed samples for the next iteration. We demonstrate the effectiveness of this iterative refinement framework via extensive experimental evaluations on the publicly available ChestXray14 dataset. AGCL achieves over 5.7\% (averaged over 14 diseases) increase in classification AUC and 7%/11% increases in Recall/Precision for the localization task compared to the state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge