Peirong Liu
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
Jun 17, 2025Abstract:We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187 subjects, aggregated from 16 publicly available sources. The dataset includes both clinical- and research-grade images, multiple MRI sequences, and a wide range of anatomical and pathological variability, including scans with large brain anomalies. Minimal preprocessing was applied to preserve the original image characteristics while reducing barriers to entry for new users. Accompanying code for self-supervised pretraining and finetuning is provided. FOMO60K is intended to support the development and benchmarking of self-supervised learning methods in medical imaging at scale.
Conditional diffusion models for guided anomaly detection in brain images using fluid-driven anomaly randomization
Jun 11, 2025Abstract:Supervised machine learning has enabled accurate pathology detection in brain MRI, but requires training data from diseased subjects that may not be readily available in some scenarios, for example, in the case of rare diseases. Reconstruction-based unsupervised anomaly detection, in particular using diffusion models, has gained popularity in the medical field as it allows for training on healthy images alone, eliminating the need for large disease-specific cohorts. These methods assume that a model trained on normal data cannot accurately represent or reconstruct anomalies. However, this assumption often fails with models failing to reconstruct healthy tissue or accurately reconstruct abnormal regions i.e., failing to remove anomalies. In this work, we introduce a novel conditional diffusion model framework for anomaly detection and healthy image reconstruction in brain MRI. Our weakly supervised approach integrates synthetically generated pseudo-pathology images into the modeling process to better guide the reconstruction of healthy images. To generate these pseudo-pathologies, we apply fluid-driven anomaly randomization to augment real pathology segmentation maps from an auxiliary dataset, ensuring that the synthetic anomalies are both realistic and anatomically coherent. We evaluate our model's ability to detect pathology, using both synthetic anomaly datasets and real pathology from the ATLAS dataset. In our extensive experiments, our model: (i) consistently outperforms variational autoencoders, and conditional and unconditional latent diffusion; and (ii) surpasses on most datasets, the performance of supervised inpainting methods with access to paired diseased/healthy images.
Scalable Segmentation for Ultra-High-Resolution Brain MR Images
May 27, 2025Abstract:Although deep learning has shown great success in 3D brain MRI segmentation, achieving accurate and efficient segmentation of ultra-high-resolution brain images remains challenging due to the lack of labeled training data for fine-scale anatomical structures and high computational demands. In this work, we propose a novel framework that leverages easily accessible, low-resolution coarse labels as spatial references and guidance, without incurring additional annotation cost. Instead of directly predicting discrete segmentation maps, our approach regresses per-class signed distance transform maps, enabling smooth, boundary-aware supervision. Furthermore, to enhance scalability, generalizability, and efficiency, we introduce a scalable class-conditional segmentation strategy, where the model learns to segment one class at a time conditioned on a class-specific input. This novel design not only reduces memory consumption during both training and testing, but also allows the model to generalize to unseen anatomical classes. We validate our method through comprehensive experiments on both synthetic and real-world datasets, demonstrating its superior performance and scalability compared to conventional segmentation approaches.
PEPSI: Pathology-Enhanced Pulse-Sequence-Invariant Representations for Brain MRI
Mar 10, 2024



Abstract:Remarkable progress has been made by data-driven machine-learning methods in the analysis of MRI scans. However, most existing MRI analysis approaches are crafted for specific MR pulse sequences (MR contrasts) and usually require nearly isotropic acquisitions. This limits their applicability to diverse real-world clinical data, where scans commonly exhibit variations in appearances due to being obtained with varying sequence parameters, resolutions, and orientations -- especially in the presence of pathology. In this paper, we propose PEPSI, the first pathology-enhanced, and pulse-sequence-invariant feature representation learning model for brain MRI. PEPSI is trained entirely on synthetic images with a novel pathology encoding strategy, and enables co-training across datasets with diverse pathologies and missing modalities. Despite variations in pathology appearances across different MR pulse sequences or the quality of acquired images (e.g., resolution, orientation, artifacts, etc), PEPSI produces a high-resolution image of reference contrast (MP-RAGE) that captures anatomy, along with an image specifically highlighting the pathology. Our experiments demonstrate PEPSI's remarkable capability for image synthesis compared with the state-of-the-art, contrast-agnostic synthesis models, as it accurately reconstructs anatomical structures while differentiating between pathology and normal tissue. We further illustrate the efficiency and effectiveness of PEPSI features for downstream pathology segmentations on five public datasets covering white matter hyperintensities and stroke lesions. Code is available at https://github.com/peirong26/PEPSI.
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI
Dec 08, 2023



Abstract:Brain atrophy and white matter hyperintensity (WMH) are critical neuroimaging features for ascertaining brain injury in cerebrovascular disease and multiple sclerosis. Automated segmentation and quantification is desirable but existing methods require high-resolution MRI with good signal-to-noise ratio (SNR). This precludes application to clinical and low-field portable MRI (pMRI) scans, thus hampering large-scale tracking of atrophy and WMH progression, especially in underserved areas where pMRI has huge potential. Here we present a method that segments white matter hyperintensity and 36 brain regions from scans of any resolution and contrast (including pMRI) without retraining. We show results on six public datasets and on a private dataset with paired high- and low-field scans (3T and 64mT), where we attain strong correlation between the WMH ($\rho$=.85) and hippocampal volumes (r=.89) estimated at both fields. Our method is publicly available as part of FreeSurfer, at: http://surfer.nmr.mgh.harvard.edu/fswiki/WMH-SynthSeg.
Brain-ID: Learning Robust Feature Representations for Brain Imaging
Nov 28, 2023Abstract:Recent learning-based approaches have made astonishing advances in calibrated medical imaging like computerized tomography, yet they struggle to generalize in uncalibrated modalities -- notoriously magnetic resonance imaging (MRI), where performance is highly sensitive to the differences in MR contrast, resolution, and orientation between the training and testing data. This prevents broad applicability to the diverse clinical acquisition protocols in the real world. We introduce Brain-ID, a robust feature representation learning strategy for brain imaging, which is contrast-agnostic, and robust to the brain anatomy of each subject regardless of the appearance of acquired images (i.e., deformation, contrast, resolution, orientation, artifacts, etc). Brain-ID is trained entirely on synthetic data, and easily adapts to downstream tasks with our proposed simple one-layer solution. We validate the robustness of Brain-ID features, and evaluate their performance in a variety of downstream applications, including both contrast-independent (anatomy reconstruction/contrast synthesis, brain segmentation), and contrast-dependent (super-resolution, bias field estimation) tasks. Extensive experiments on 6 public datasets demonstrate that Brain-ID achieves state-of-the-art performance in all tasks, and more importantly, preserves its performance when only limited training data is available.
A Unified Model for Tracking and Image-Video Detection Has More Power
Nov 20, 2022



Abstract:Objection detection (OD) has been one of the most fundamental tasks in computer vision. Recent developments in deep learning have pushed the performance of image OD to new heights by learning-based, data-driven approaches. On the other hand, video OD remains less explored, mostly due to much more expensive data annotation needs. At the same time, multi-object tracking (MOT) which requires reasoning about track identities and spatio-temporal trajectories, shares similar spirits with video OD. However, most MOT datasets are class-specific (e.g., person-annotated only), which constrains a model's flexibility to perform tracking on other objects. We propose TrIVD (Tracking and Image-Video Detection), the first framework that unifies image OD, video OD, and MOT within one end-to-end model. To handle the discrepancies and semantic overlaps across datasets, TrIVD formulates detection/tracking as grounding and reasons about object categories via visual-text alignments. The unified formulation enables cross-dataset, multi-task training, and thus equips TrIVD with the ability to leverage frame-level features, video-level spatio-temporal relations, as well as track identity associations. With such joint training, we can now extend the knowledge from OD data, that comes with much richer object category annotations, to MOT and achieve zero-shot tracking capability. Experiments demonstrate that TrIVD achieves state-of-the-art performances across all image/video OD and MOT tasks.
Efficient conditioned face animation using frontally-viewed embedding
Mar 16, 2022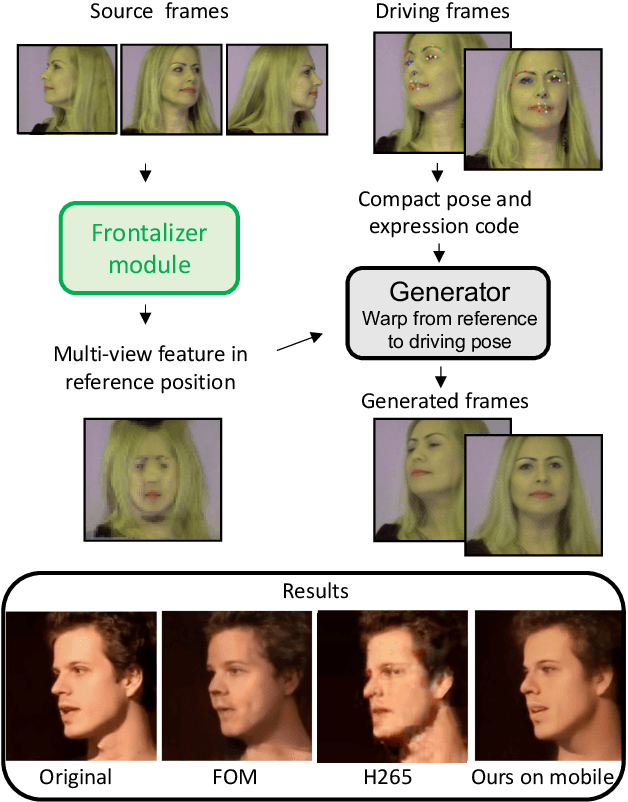

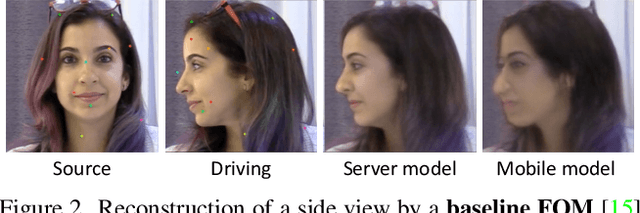
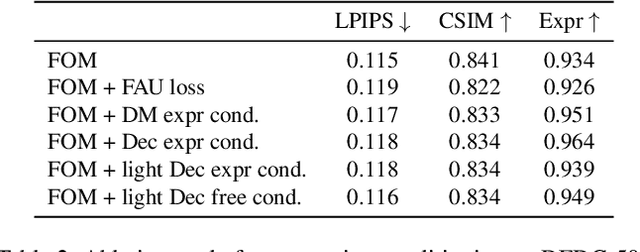
Abstract:As the quality of few shot facial animation from landmarks increases, new applications become possible, such as ultra low bandwidth video chat compression with a high degree of realism. However, there are some important challenges to tackle in order to improve the experience in real world conditions. In particular, the current approaches fail to represent profile views without distortions, while running in a low compute regime. We focus on this key problem by introducing a multi-frames embedding dubbed Frontalizer to improve profile views rendering. In addition to this core improvement, we explore the learning of a latent code conditioning generations along with landmarks to better convey facial expressions. Our dense models achieves 22% of improvement in perceptual quality and 73% reduction of landmark error over the first order model baseline on a subset of DFDC videos containing head movements. Declined with mobile architectures, our models outperform the previous state-of-the-art (improving perceptual quality by more than 16% and reducing landmark error by more than 47% on two datasets) while running on real time on iPhone 8 with very low bandwidth requirements.
Fluid registration between lung CT and stationary chest tomosynthesis images
Mar 06, 2022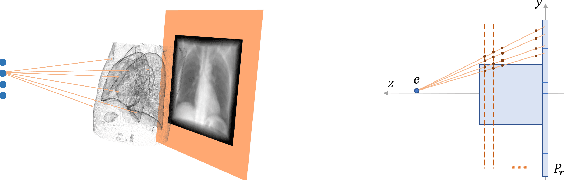
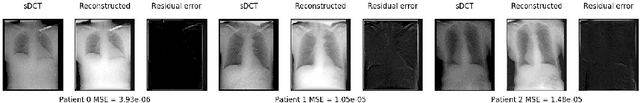
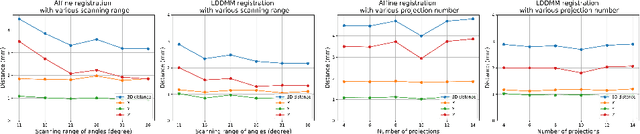
Abstract:Registration is widely used in image-guided therapy and image-guided surgery to estimate spatial correspondences between organs of interest between planning and treatment images. However, while high-quality computed tomography (CT) images are often available at planning time, limited angle acquisitions are frequently used during treatment because of radiation concerns or imaging time constraints. This requires algorithms to register CT images based on limited angle acquisitions. We, therefore, formulate a 3D/2D registration approach which infers a 3D deformation based on measured projections and digitally reconstructed radiographs of the CT. Most 3D/2D registration approaches use simple transformation models or require complex mathematical derivations to formulate the underlying optimization problem. Instead, our approach entirely relies on differentiable operations which can be combined with modern computational toolboxes supporting automatic differentiation. This then allows for rapid prototyping, integration with deep neural networks, and to support a variety of transformation models including fluid flow models. We demonstrate our approach for the registration between CT and stationary chest tomosynthesis (sDCT) images and show how it naturally leads to an iterative image reconstruction approach.
Deep Decomposition for Stochastic Normal-Abnormal Transport
Nov 29, 2021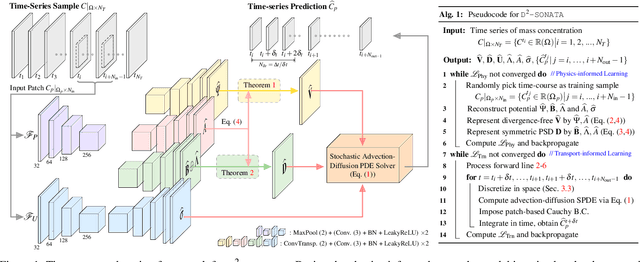
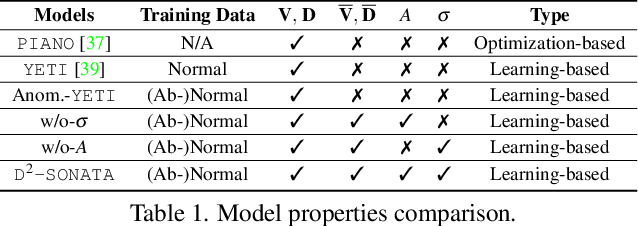
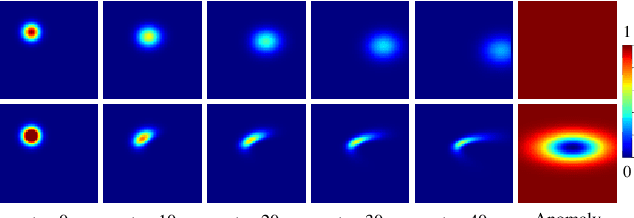
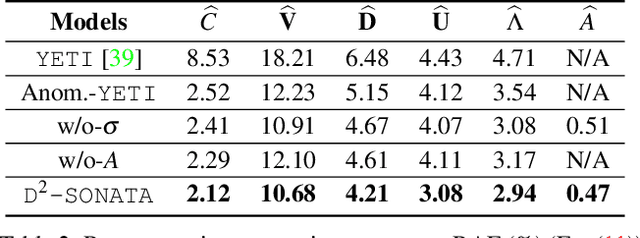
Abstract:Advection-diffusion equations describe a large family of natural transport processes, e.g., fluid flow, heat transfer, and wind transport. They are also used for optical flow and perfusion imaging computations. We develop a machine learning model, D^2-SONATA, built upon a stochastic advection-diffusion equation, which predicts the velocity and diffusion fields that drive 2D/3D image time-series of transport. In particular, our proposed model incorporates a model of transport atypicality, which isolates abnormal differences between expected normal transport behavior and the observed transport. In a medical context such a normal-abnormal decomposition can be used, for example, to quantify pathologies. Specifically, our model identifies the advection and diffusion contributions from the transport time-series and simultaneously predicts an anomaly value field to provide a decomposition into normal and abnormal advection and diffusion behavior. To achieve improved estimation performance for the velocity and diffusion-tensor fields underlying the advection-diffusion process and for the estimation of the anomaly fields, we create a 2D/3D anomaly-encoded advection-diffusion simulator, which allows for supervised learning. We further apply our model on a brain perfusion dataset from ischemic stroke patients via transfer learning. Extensive comparisons demonstrate that our model successfully distinguishes stroke lesions (abnormal) from normal brain regions, while reconstructing the underlying velocity and diffusion tensor fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge