Mostafa Mehdipour Ghazi
Deep Learning-Based Regional White Matter Hyperintensity Mapping as a Robust Biomarker for Alzheimer's Disease
Nov 18, 2025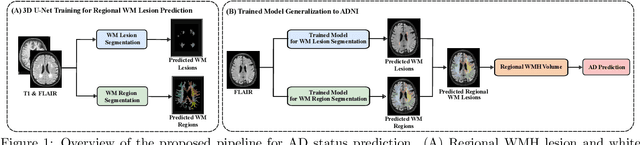
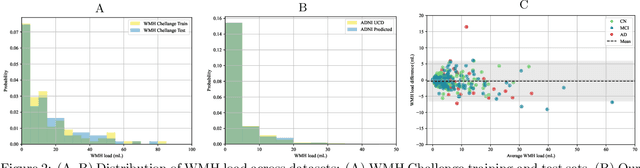
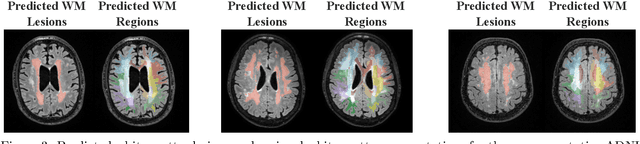
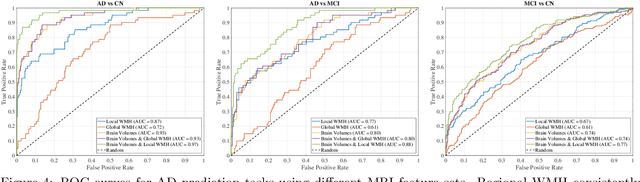
Abstract:White matter hyperintensities (WMH) are key imaging markers in cognitive aging, Alzheimer's disease (AD), and related dementias. Although automated methods for WMH segmentation have advanced, most provide only global lesion load and overlook their spatial distribution across distinct white matter regions. We propose a deep learning framework for robust WMH segmentation and localization, evaluated across public datasets and an independent Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. Our results show that the predicted lesion loads are in line with the reference WMH estimates, confirming the robustness to variations in lesion load, acquisition, and demographics. Beyond accurate segmentation, we quantify WMH load within anatomically defined regions and combine these measures with brain structure volumes to assess diagnostic value. Regional WMH volumes consistently outperform global lesion burden for disease classification, and integration with brain atrophy metrics further improves performance, reaching area under the curve (AUC) values up to 0.97. Several spatially distinct regions, particularly within anterior white matter tracts, are reproducibly associated with diagnostic status, indicating localized vulnerability in AD. These results highlight the added value of regional WMH quantification. Incorporating localized lesion metrics alongside atrophy markers may enhance early diagnosis and stratification in neurodegenerative disorders.
MRI Embeddings Complement Clinical Predictors for Cognitive Decline Modeling in Alzheimer's Disease Cohorts
Nov 18, 2025Abstract:Accurate modeling of cognitive decline in Alzheimer's disease is essential for early stratification and personalized management. While tabular predictors provide robust markers of global risk, their ability to capture subtle brain changes remains limited. In this study, we evaluate the predictive contributions of tabular and imaging-based representations, with a focus on transformer-derived Magnetic Resonance Imaging (MRI) embeddings. We introduce a trajectory-aware labeling strategy based on Dynamic Time Warping clustering to capture heterogeneous patterns of cognitive change, and train a 3D Vision Transformer (ViT) via unsupervised reconstruction on harmonized and augmented MRI data to obtain anatomy-preserving embeddings without progression labels. The pretrained encoder embeddings are subsequently assessed using both traditional machine learning classifiers and deep learning heads, and compared against tabular representations and convolutional network baselines. Results highlight complementary strengths across modalities. Clinical and volumetric features achieved the highest AUCs of around 0.70 for predicting mild and severe progression, underscoring their utility in capturing global decline trajectories. In contrast, MRI embeddings from the ViT model were most effective in distinguishing cognitively stable individuals with an AUC of 0.71. However, all approaches struggled in the heterogeneous moderate group. These findings indicate that clinical features excel in identifying high-risk extremes, whereas transformer-based MRI embeddings are more sensitive to subtle markers of stability, motivating multimodal fusion strategies for AD progression modeling.
Tooth-Diffusion: Guided 3D CBCT Synthesis with Fine-Grained Tooth Conditioning
Aug 19, 2025


Abstract:Despite the growing importance of dental CBCT scans for diagnosis and treatment planning, generating anatomically realistic scans with fine-grained control remains a challenge in medical image synthesis. In this work, we propose a novel conditional diffusion framework for 3D dental volume generation, guided by tooth-level binary attributes that allow precise control over tooth presence and configuration. Our approach integrates wavelet-based denoising diffusion, FiLM conditioning, and masked loss functions to focus learning on relevant anatomical structures. We evaluate the model across diverse tasks, such as tooth addition, removal, and full dentition synthesis, using both paired and distributional similarity metrics. Results show strong fidelity and generalization with low FID scores, robust inpainting performance, and SSIM values above 0.91 even on unseen scans. By enabling realistic, localized modification of dentition without rescanning, this work opens opportunities for surgical planning, patient communication, and targeted data augmentation in dental AI workflows. The codes are available at: https://github.com/djafar1/tooth-diffusion.
A large-scale heterogeneous 3D magnetic resonance brain imaging dataset for self-supervised learning
Jun 17, 2025Abstract:We present FOMO60K, a large-scale, heterogeneous dataset of 60,529 brain Magnetic Resonance Imaging (MRI) scans from 13,900 sessions and 11,187 subjects, aggregated from 16 publicly available sources. The dataset includes both clinical- and research-grade images, multiple MRI sequences, and a wide range of anatomical and pathological variability, including scans with large brain anomalies. Minimal preprocessing was applied to preserve the original image characteristics while reducing barriers to entry for new users. Accompanying code for self-supervised pretraining and finetuning is provided. FOMO60K is intended to support the development and benchmarking of self-supervised learning methods in medical imaging at scale.
Assessing the Efficacy of Classical and Deep Neuroimaging Biomarkers in Early Alzheimer's Disease Diagnosis
Oct 31, 2024



Abstract:Alzheimer's disease (AD) is the leading cause of dementia, and its early detection is crucial for effective intervention, yet current diagnostic methods often fall short in sensitivity and specificity. This study aims to detect significant indicators of early AD by extracting and integrating various imaging biomarkers, including radiomics, hippocampal texture descriptors, cortical thickness measurements, and deep learning features. We analyze structural magnetic resonance imaging (MRI) scans from the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohorts, utilizing comprehensive image analysis and machine learning techniques. Our results show that combining multiple biomarkers significantly improves detection accuracy. Radiomics and texture features emerged as the most effective predictors for early AD, achieving AUCs of 0.88 and 0.72 for AD and MCI detection, respectively. Although deep learning features proved to be less effective than traditional approaches, incorporating age with other biomarkers notably enhanced MCI detection performance. Additionally, our findings emphasize the continued importance of classical imaging biomarkers in the face of modern deep-learning approaches, providing a robust framework for early AD diagnosis.
Yucca: A Deep Learning Framework For Medical Image Analysis
Jul 29, 2024


Abstract:Medical image analysis using deep learning frameworks has advanced healthcare by automating complex tasks, but many existing frameworks lack flexibility, modularity, and user-friendliness. To address these challenges, we introduce Yucca, an open-source AI framework available at https://github.com/Sllambias/yucca, designed specifically for medical imaging applications and built on PyTorch and PyTorch Lightning. Yucca features a three-tiered architecture: Functional, Modules, and Pipeline, providing a comprehensive and customizable solution. Evaluated across diverse tasks such as cerebral microbleeds detection, white matter hyperintensity segmentation, and hippocampus segmentation, Yucca achieves state-of-the-art results, demonstrating its robustness and versatility. Yucca offers a powerful, flexible, and user-friendly platform for medical image analysis, inviting community contributions to advance its capabilities and impact.
Non-Reference Quality Assessment for Medical Imaging: Application to Synthetic Brain MRIs
Jul 20, 2024



Abstract:Generating high-quality synthetic data is crucial for addressing challenges in medical imaging, such as domain adaptation, data scarcity, and privacy concerns. Existing image quality metrics often rely on reference images, are tailored for group comparisons, or are intended for 2D natural images, limiting their efficacy in complex domains like medical imaging. This study introduces a novel deep learning-based non-reference approach to assess brain MRI quality by training a 3D ResNet. The network is designed to estimate quality across six distinct artifacts commonly encountered in MRI scans. Additionally, a diffusion model is trained on diverse datasets to generate synthetic 3D images of high fidelity. The approach leverages several datasets for training and comprehensive quality assessment, benchmarking against state-of-the-art metrics for real and synthetic images. Results demonstrate superior performance in accurately estimating distortions and reflecting image quality from multiple perspectives. Notably, the method operates without reference images, indicating its applicability for evaluating deep generative models. Besides, the quality scores in the [0, 1] range provide an intuitive assessment of image quality across heterogeneous datasets. Evaluation of generated images offers detailed insights into specific artifacts, guiding strategies for improving generative models to produce high-quality synthetic images. This study presents the first comprehensive method for assessing the quality of real and synthetic 3D medical images in MRI contexts without reliance on reference images.
Data Augmentation-Based Unsupervised Domain Adaptation In Medical Imaging
Aug 08, 2023Abstract:Deep learning-based models in medical imaging often struggle to generalize effectively to new scans due to data heterogeneity arising from differences in hardware, acquisition parameters, population, and artifacts. This limitation presents a significant challenge in adopting machine learning models for clinical practice. We propose an unsupervised method for robust domain adaptation in brain MRI segmentation by leveraging MRI-specific augmentation techniques. To evaluate the effectiveness of our method, we conduct extensive experiments across diverse datasets, modalities, and segmentation tasks, comparing against the state-of-the-art methods. The results show that our proposed approach achieves high accuracy, exhibits broad applicability, and showcases remarkable robustness against domain shift in various tasks, surpassing the state-of-the-art performance in the majority of cases.
Deep Learning-Based Assessment of Cerebral Microbleeds in COVID-19
Jan 23, 2023Abstract:Cerebral Microbleeds (CMBs), typically captured as hypointensities from susceptibility-weighted imaging (SWI), are particularly important for the study of dementia, cerebrovascular disease, and normal aging. Recent studies on COVID-19 have shown an increase in CMBs of coronavirus cases. Automatic detection of CMBs is challenging due to the small size and amount of CMBs making the classes highly imbalanced, lack of publicly available annotated data, and similarity with CMB mimics such as calcifications, irons, and veins. Hence, the existing deep learning methods are mostly trained on very limited research data and fail to generalize to unseen data with high variability and cannot be used in clinical setups. To this end, we propose an efficient 3D deep learning framework that is actively trained on multi-domain data. Two public datasets assigned for normal aging, stroke, and Alzheimer's disease analysis as well as an in-house dataset for COVID-19 assessment are used to train and evaluate the models. The obtained results show that the proposed method is robust to low-resolution images and achieves 78% recall and 80% precision on the entire test set with an average false positive of 1.6 per scan.
FAST-AID Brain: Fast and Accurate Segmentation Tool using Artificial Intelligence Developed for Brain
Aug 30, 2022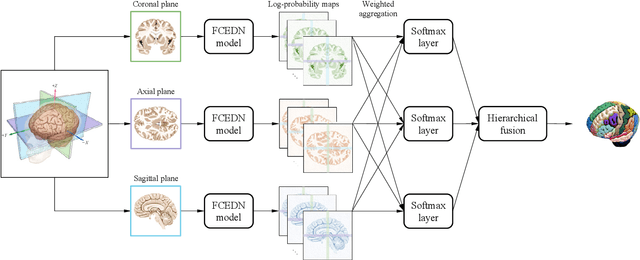

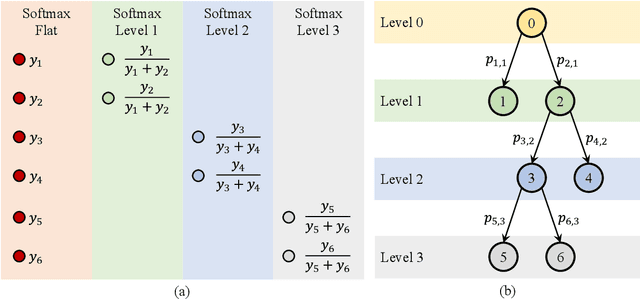
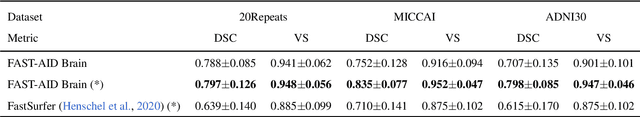
Abstract:Medical images used in clinical practice are heterogeneous and not the same quality as scans studied in academic research. Preprocessing breaks down in extreme cases when anatomy, artifacts, or imaging parameters are unusual or protocols are different. Methods robust to these variations are most needed. A novel deep learning method is proposed for fast and accurate segmentation of the human brain into 132 regions. The proposed model uses an efficient U-Net-like network and benefits from the intersection points of different views and hierarchical relations for the fusion of the orthogonal 2D planes and brain labels during the end-to-end training. Weakly supervised learning is deployed to take the advantage of partially labeled data for the whole brain segmentation and estimation of the intracranial volume (ICV). Moreover, data augmentation is used to expand the magnetic resonance imaging (MRI) data by generating realistic brain scans with high variability for robust training of the model while preserving data privacy. The proposed method can be applied to brain MRI data including skull or any other artifacts without preprocessing the images or a drop in performance. Several experiments using different atlases are conducted to evaluate the segmentation performance of the trained model compared to the state-of-the-art, and the results show higher segmentation accuracy and robustness of the proposed model compared to the existing methods across different intra- and inter-domain datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge