Yueh Z. Lee
R2Gen-Mamba: A Selective State Space Model for Radiology Report Generation
Oct 21, 2024Abstract:Radiology report generation is crucial in medical imaging,but the manual annotation process by physicians is time-consuming and labor-intensive, necessitating the develop-ment of automatic report generation methods. Existingresearch predominantly utilizes Transformers to generateradiology reports, which can be computationally intensive,limiting their use in real applications. In this work, we presentR2Gen-Mamba, a novel automatic radiology report genera-tion method that leverages the efficient sequence processingof the Mamba with the contextual benefits of Transformerarchitectures. Due to lower computational complexity ofMamba, R2Gen-Mamba not only enhances training and in-ference efficiency but also produces high-quality reports.Experimental results on two benchmark datasets with morethan 210,000 X-ray image-report pairs demonstrate the ef-fectiveness of R2Gen-Mamba regarding report quality andcomputational efficiency compared with several state-of-the-art methods. The source code can be accessed online.
Autonomous Medical Needle Steering In Vivo
Nov 04, 2022Abstract:The use of needles to access sites within organs is fundamental to many interventional medical procedures both for diagnosis and treatment. Safe and accurate navigation of a needle through living tissue to an intra-tissue target is currently often challenging or infeasible due to the presence of anatomical obstacles in the tissue, high levels of uncertainty, and natural tissue motion (e.g., due to breathing). Medical robots capable of automating needle-based procedures in vivo have the potential to overcome these challenges and enable an enhanced level of patient care and safety. In this paper, we show the first medical robot that autonomously navigates a needle inside living tissue around anatomical obstacles to an intra-tissue target. Our system leverages an aiming device and a laser-patterned highly flexible steerable needle, a type of needle capable of maneuvering along curvilinear trajectories to avoid obstacles. The autonomous robot accounts for anatomical obstacles and uncertainty in living tissue/needle interaction with replanning and control and accounts for respiratory motion by defining safe insertion time windows during the breathing cycle. We apply the system to lung biopsy, which is critical in the diagnosis of lung cancer, the leading cause of cancer-related death in the United States. We demonstrate successful performance of our system in multiple in vivo porcine studies and also demonstrate that our approach leveraging autonomous needle steering outperforms a standard manual clinical technique for lung nodule access.
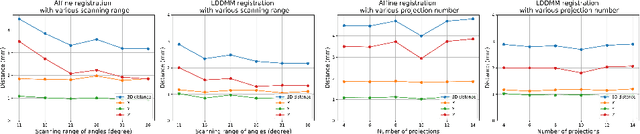
LiftReg: Limited Angle 2D/3D Deformable Registration
Mar 10, 2022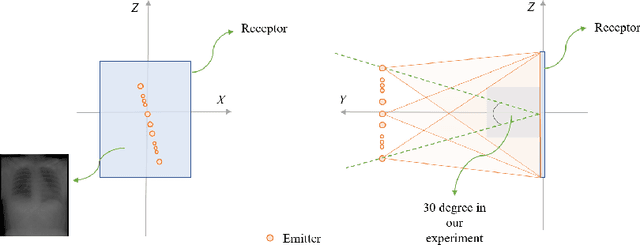
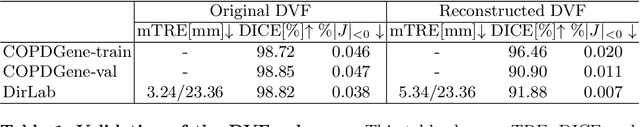
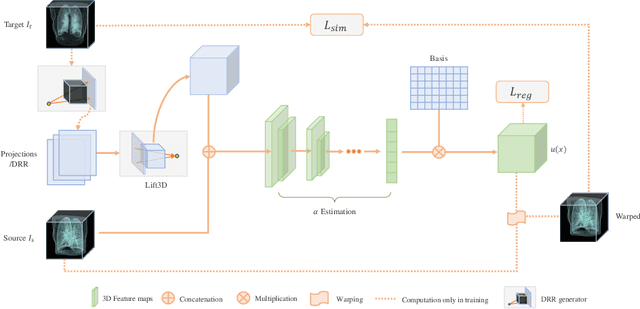
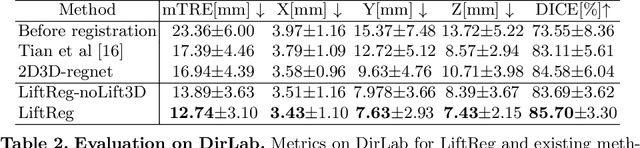
Abstract:We propose LiftReg, a 2D/3D deformable registration approach. LiftReg is a deep registration framework which is trained using sets of digitally reconstructed radiographs (DRR) and computed tomography (CT) image pairs. By using simulated training data, LiftReg can use a high-quality CT-CT image similarity measure, which helps the network to learn a high-quality deformation space. To further improve registration quality and to address the inherent depth ambiguities of very limited angle acquisitions, we propose to use features extracted from the backprojected 2D images and a statistical deformation model. We test our approach on the DirLab lung registration dataset and show that it outperforms an existing learning-based pairwise registration approach.
Fluid registration between lung CT and stationary chest tomosynthesis images
Mar 06, 2022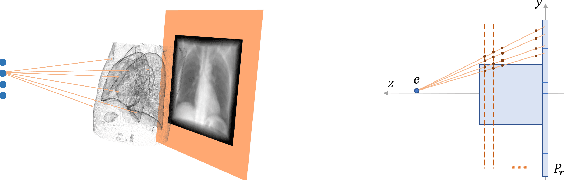
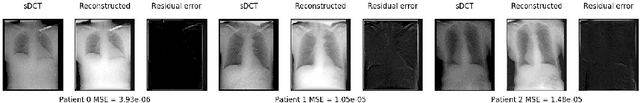
Abstract:Registration is widely used in image-guided therapy and image-guided surgery to estimate spatial correspondences between organs of interest between planning and treatment images. However, while high-quality computed tomography (CT) images are often available at planning time, limited angle acquisitions are frequently used during treatment because of radiation concerns or imaging time constraints. This requires algorithms to register CT images based on limited angle acquisitions. We, therefore, formulate a 3D/2D registration approach which infers a 3D deformation based on measured projections and digitally reconstructed radiographs of the CT. Most 3D/2D registration approaches use simple transformation models or require complex mathematical derivations to formulate the underlying optimization problem. Instead, our approach entirely relies on differentiable operations which can be combined with modern computational toolboxes supporting automatic differentiation. This then allows for rapid prototyping, integration with deep neural networks, and to support a variety of transformation models including fluid flow models. We demonstrate our approach for the registration between CT and stationary chest tomosynthesis (sDCT) images and show how it naturally leads to an iterative image reconstruction approach.
Discovering Hidden Physics Behind Transport Dynamics
Nov 24, 2020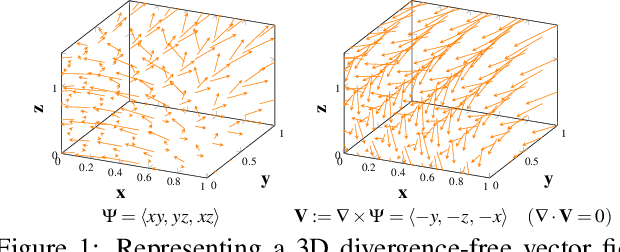
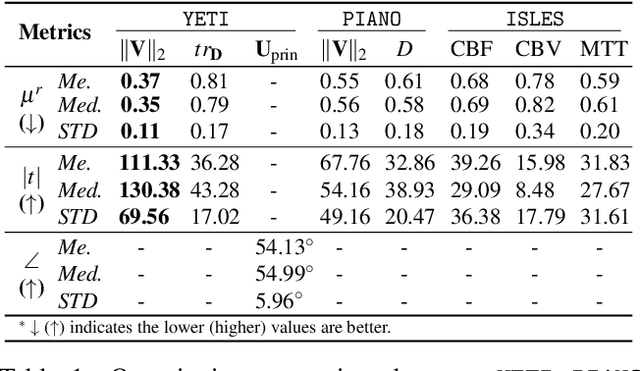
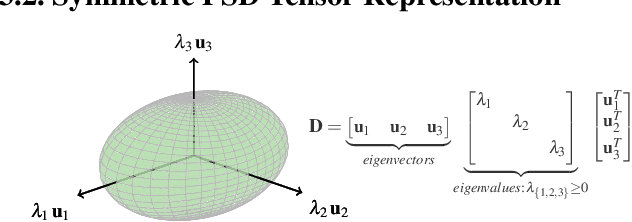
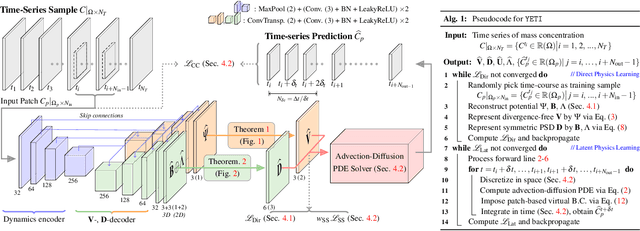
Abstract:Transport processes are ubiquitous. They are, for example, at the heart of optical flow approaches; or of perfusion imaging, where blood transport is assessed, most commonly by injecting a tracer. An advection-diffusion equation is widely used to describe these transport phenomena. Our goal is estimating the underlying physics of advection-diffusion equations, expressed as velocity and diffusion tensor fields. We propose a learning framework (YETI) building on an auto-encoder structure between 2D and 3D image time-series, which incorporates the advection-diffusion model. To help with identifiability, we develop an advection-diffusion simulator which allows pre-training of our model by supervised learning using the velocity and diffusion tensor fields. Instead of directly learning these velocity and diffusion tensor fields, we introduce representations that assure incompressible flow and symmetric positive semi-definite diffusion fields and demonstrate the additional benefits of these representations on improving estimation accuracy. We further use transfer learning to apply YETI on a public brain magnetic resonance (MR) perfusion dataset of stroke patients and show its ability to successfully distinguish stroke lesions from normal brain regions via the estimated velocity and diffusion tensor fields.
Perfusion Imaging: A Data Assimilation Approach
Sep 06, 2020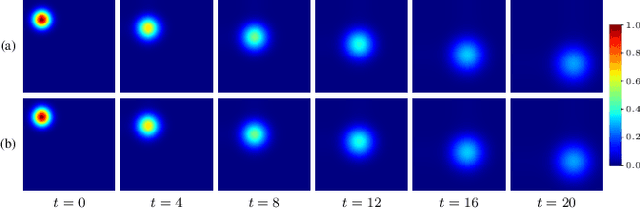
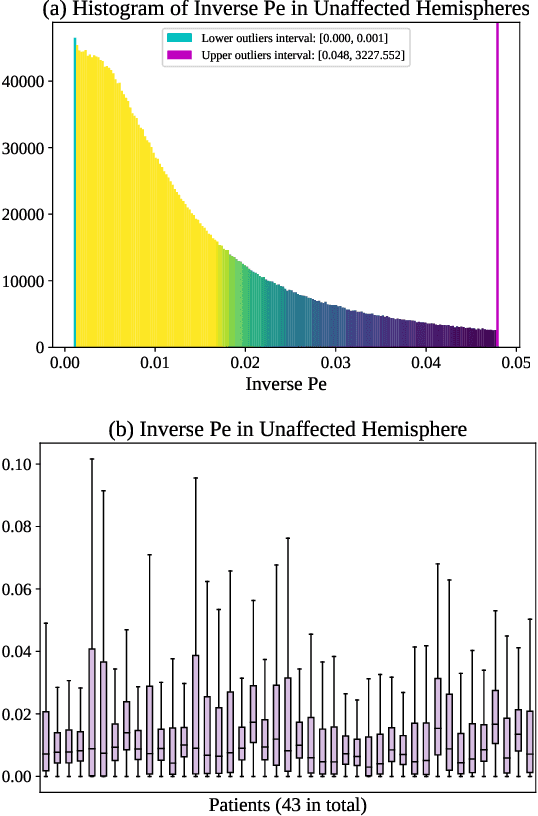
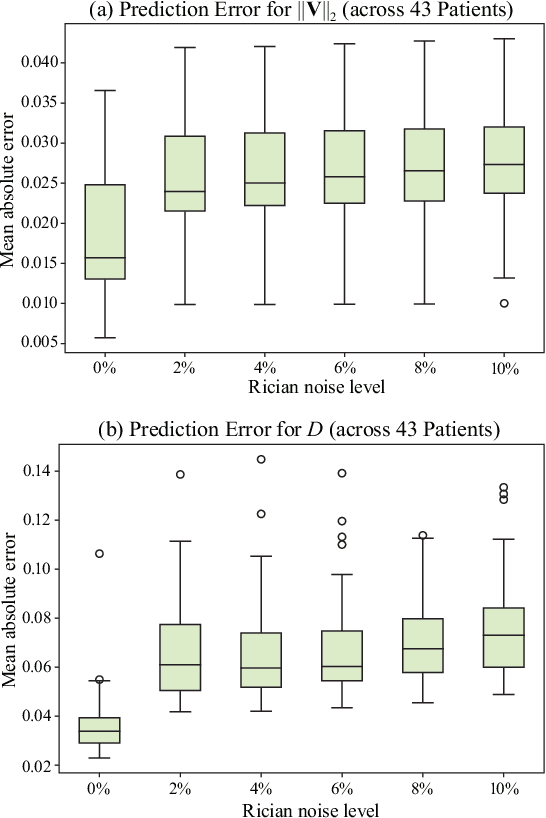
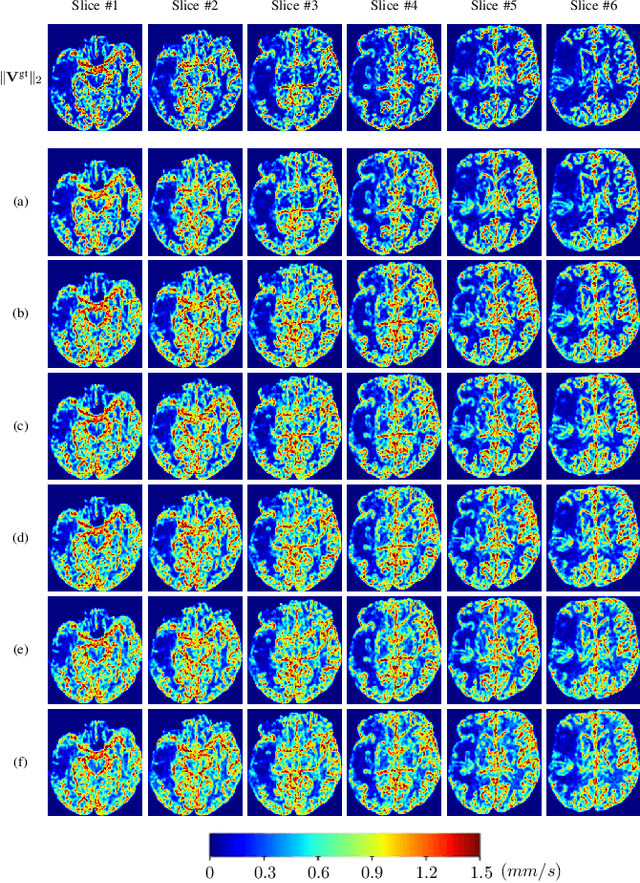
Abstract:Perfusion imaging (PI) is clinically used to assess strokes and brain tumors. Commonly used PI approaches based on magnetic resonance imaging (MRI) or computed tomography (CT) measure the effect of a contrast agent moving through blood vessels and into tissue. Contrast-agent free approaches, for example, based on intravoxel incoherent motion, also exist, but are so far not routinely used clinically. These methods rely on estimating on the arterial input function (AIF) to approximately model tissue perfusion, neglecting spatial dependencies, and reliably estimating the AIF is also non-trivial, leading to difficulties with standardizing perfusion measures. In this work we therefore propose a data-assimilation approach (PIANO) which estimates the velocity and diffusion fields of an advection-diffusion model that best explains the contrast dynamics. PIANO accounts for spatial dependencies and neither requires estimating the AIF nor relies on a particular contrast agent bolus shape. Specifically, we propose a convenient parameterization of the estimation problem, a numerical estimation approach, and extensively evaluate PIANO. We demonstrate that PIANO can successfully resolve velocity and diffusion field ambiguities and results in sensitive measures for the assessment of stroke, comparing favorably to conventional measures of perfusion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge