Markus Rempfler
The MICCAI Hackathon on reproducibility, diversity, and selection of papers at the MICCAI conference
Mar 04, 2021
Abstract:The MICCAI conference has encountered tremendous growth over the last years in terms of the size of the community, as well as the number of contributions and their technical success. With this growth, however, come new challenges for the community. Methods are more difficult to reproduce and the ever-increasing number of paper submissions to the MICCAI conference poses new questions regarding the selection process and the diversity of topics. To exchange, discuss, and find novel and creative solutions to these challenges, a new format of a hackathon was initiated as a satellite event at the MICCAI 2020 conference: The MICCAI Hackathon. The first edition of the MICCAI Hackathon covered the topics reproducibility, diversity, and selection of MICCAI papers. In the manner of a small think-tank, participants collaborated to find solutions to these challenges. In this report, we summarize the insights from the MICCAI Hackathon into immediate and long-term measures to address these challenges. The proposed measures can be seen as starting points and guidelines for discussions and actions to possibly improve the MICCAI conference with regards to reproducibility, diversity, and selection of papers.
RDCNet: Instance segmentation with a minimalist recurrent residual network
Oct 02, 2020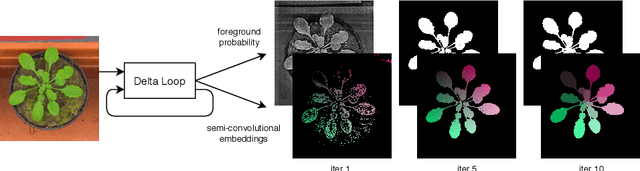
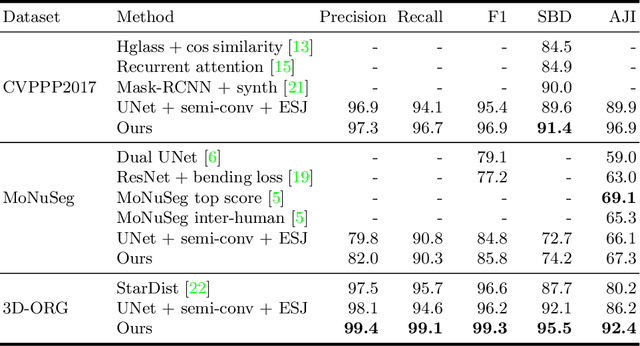
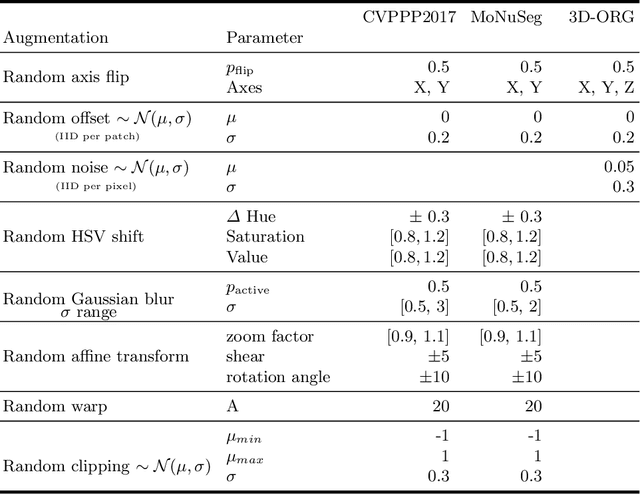
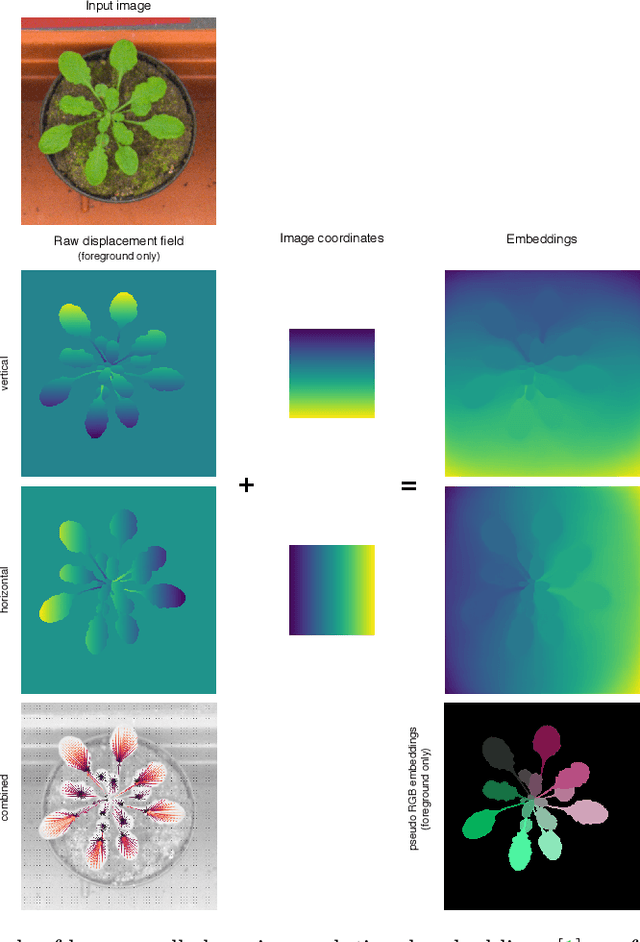
Abstract:Instance segmentation is a key step for quantitative microscopy. While several machine learning based methods have been proposed for this problem, most of them rely on computationally complex models that are trained on surrogate tasks. Building on recent developments towards end-to-end trainable instance segmentation, we propose a minimalist recurrent network called recurrent dilated convolutional network (RDCNet), consisting of a shared stacked dilated convolution (sSDC) layer that iteratively refines its output and thereby generates interpretable intermediate predictions. It is light-weight and has few critical hyperparameters, which can be related to physical aspects such as object size or density.We perform a sensitivity analysis of its main parameters and we demonstrate its versatility on 3 tasks with different imaging modalities: nuclear segmentation of H&E slides, of 3D anisotropic stacks from light-sheet fluorescence microscopy and leaf segmentation of top-view images of plants. It achieves state-of-the-art on 2 of the 3 datasets.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020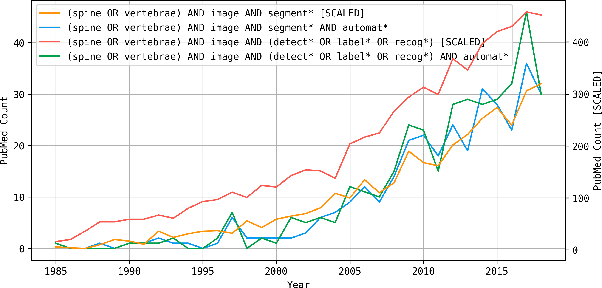
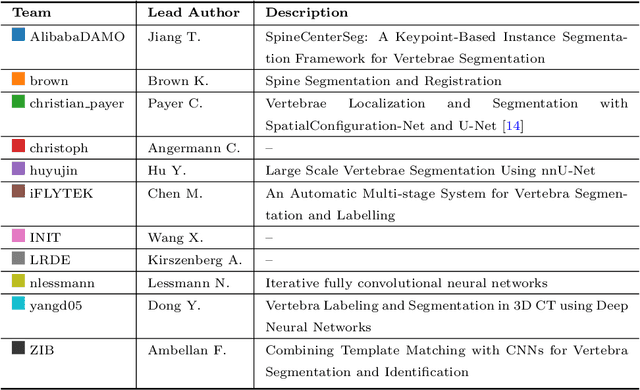
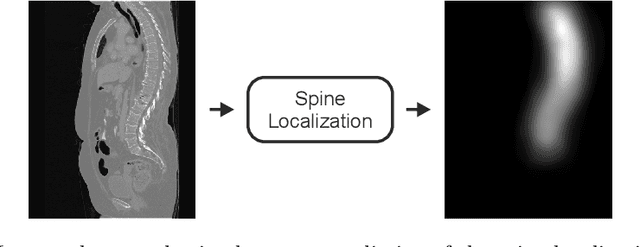
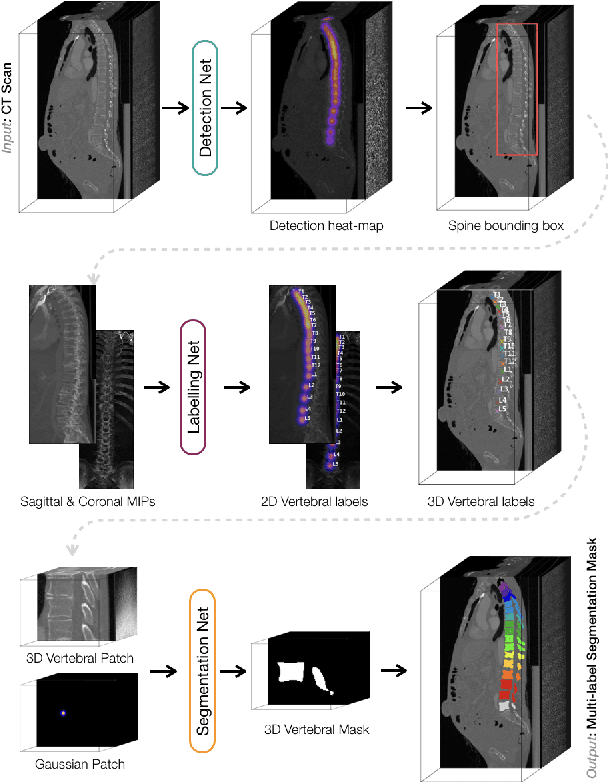
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
Probabilistic Point Cloud Reconstructions for Vertebral Shape Analysis
Aug 02, 2019
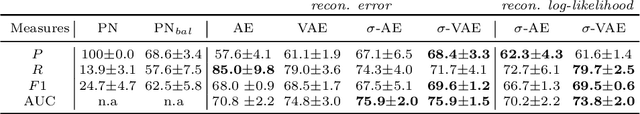
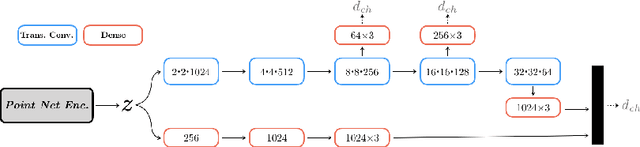
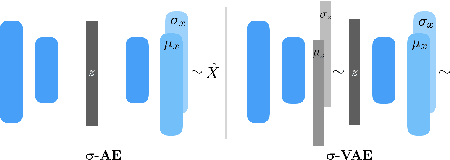
Abstract:We propose an auto-encoding network architecture for point clouds (PC) capable of extracting shape signatures without supervision. Building on this, we (i) design a loss function capable of modelling data variance on PCs which are unstructured, and (ii) regularise the latent space as in a variational auto-encoder, both of which increase the auto-encoders' descriptive capacity while making them probabilistic. Evaluating the reconstruction quality of our architectures, we employ them for detecting vertebral fractures without any supervision. By learning to efficiently reconstruct only healthy vertebrae, fractures are detected as anomalous reconstructions. Evaluating on a dataset containing $\sim$1500 vertebrae, we achieve area-under-ROC curve of $>$75%, without using intensity-based features.
Adversarially Learning a Local Anatomical Prior: Vertebrae Labelling with 2D reformations
Mar 03, 2019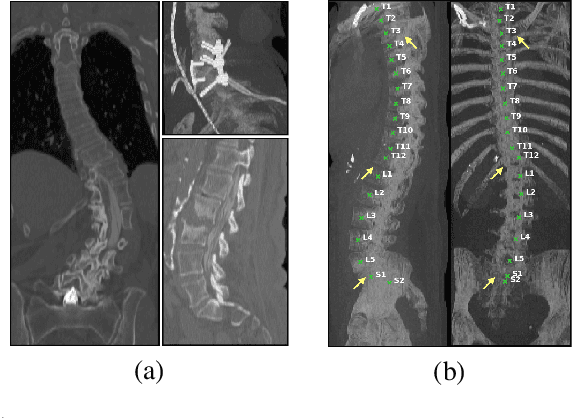
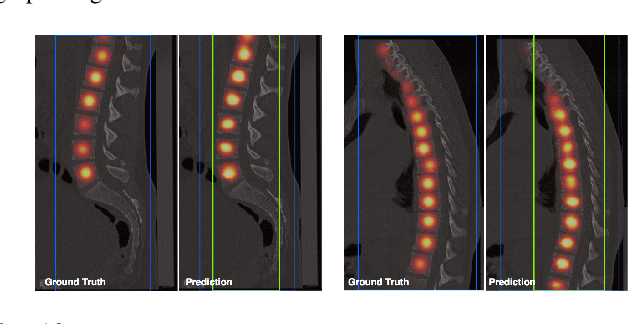
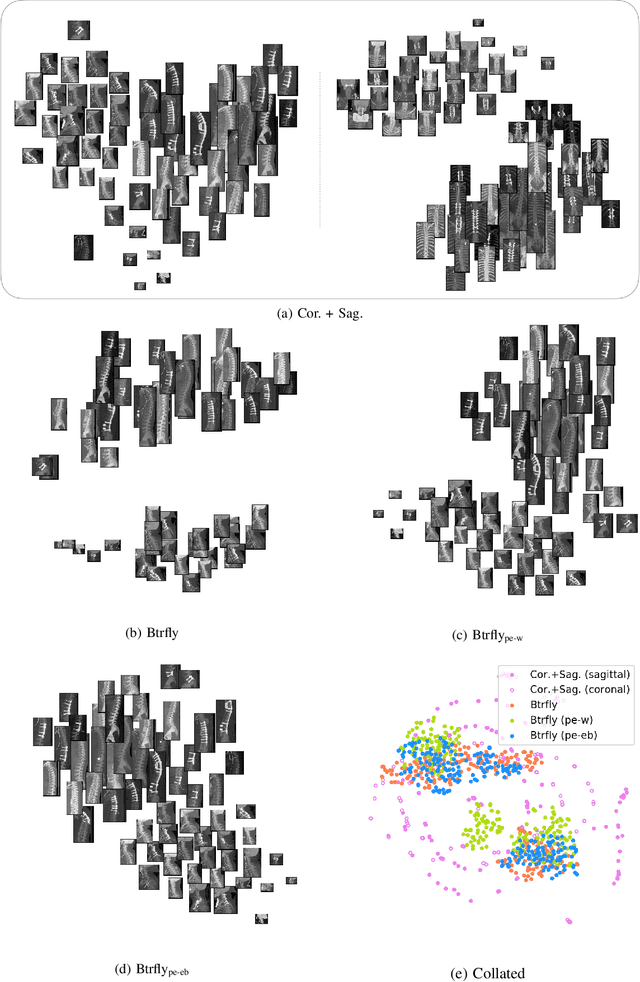
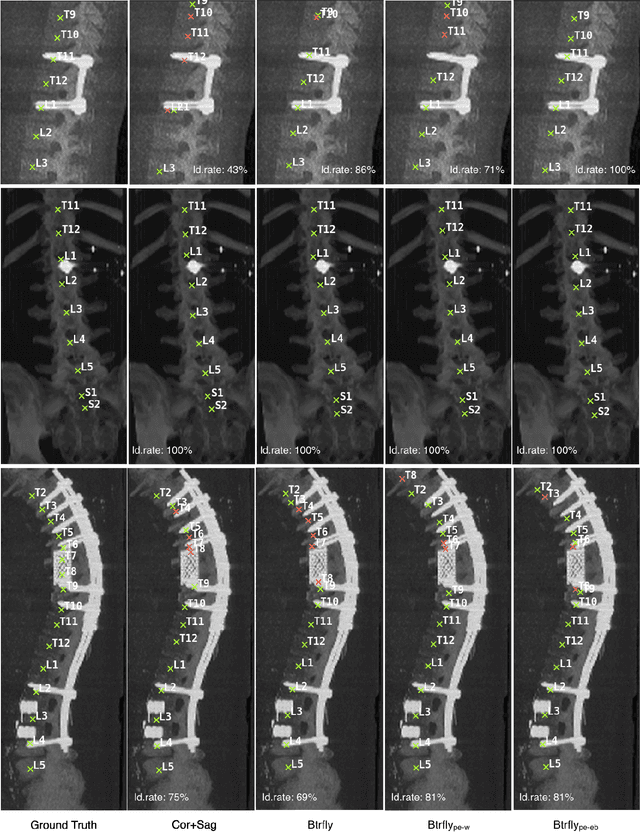
Abstract:Robust localisation and identification of vertebrae, jointly termed vertebrae labelling, in computed tomography (CT) images is an essential component of automated spine analysis. Current approaches for this task mostly work with 3D scans and are comprised of a sequence of multiple networks. Contrarily, our approach relies only on 2D reformations, enabling us to design an end-to-end trainable, standalone network. Our contribution includes: (1) Inspired by the workflow of human experts, a novel butterfly-shaped network architecture (termed Btrfly net) that efficiently combines information across sufficiently-informative sagittal and coronal reformations. (2) Two adversarial training regimes that encode an anatomical prior of the spine's shape into the Btrfly net, each enforcing the prior in a distinct manner. We evaluate our approach on a public benchmarking dataset of 302 CT scans achieving a performance comparable to state-of-art methods (identification rate of $>$88%) without any post-processing stages. Addressing its translation to clinical settings, an in-house dataset of 65 CT scans with a higher data variability is introduced, where we discuss refinements that render our approach robust to such scenarios.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019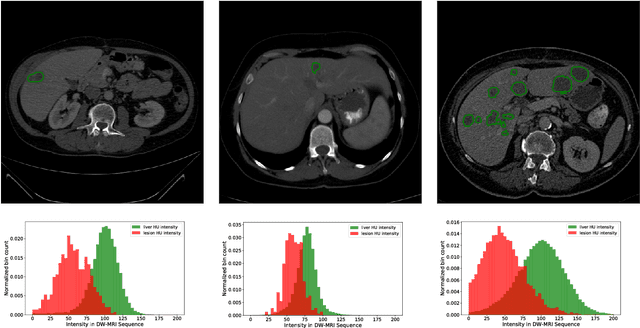
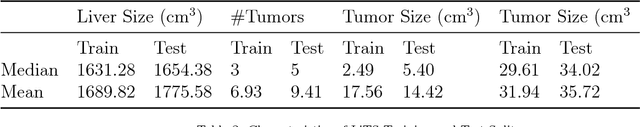
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Btrfly Net: Vertebrae Labelling with Energy-based Adversarial Learning of Local Spine Prior
Apr 04, 2018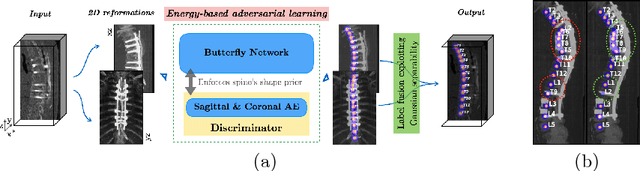
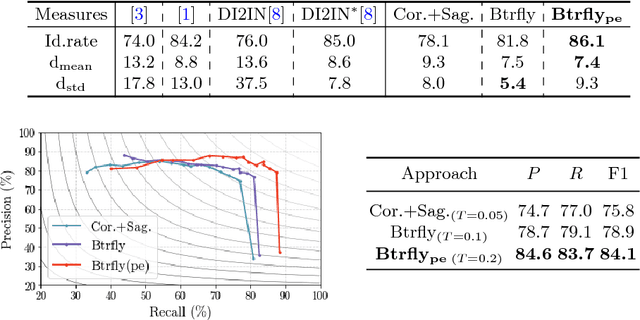
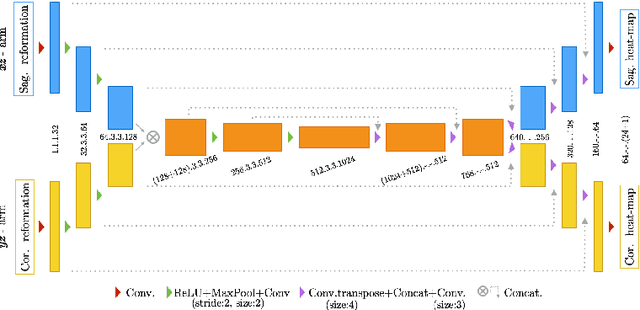
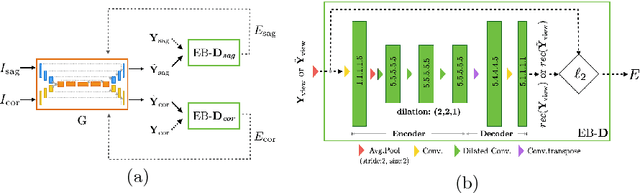
Abstract:Robust localisation and identification of vertebrae is an essential part of automated spine analysis. The contribution of this work to the task is two-fold: (1) Inspired by the human expert, we hypothesise that a sagittal and coronal reformation of the spine contain sufficient information for labelling the vertebrae. Thereby, we propose a butterfly-shaped network architecture (termed Btrfly Net) that efficiently combines the information across the reformations. (2) Underpinning the Btrfly net, we present an energy-based adversarial training regime that encodes the local spine structure as an anatomical prior into the network, thereby enabling it to achieve state-of-art performance in all standard metrics on a benchmark dataset of 302 scans without any post-processing during inference.
Efficient Algorithms for Moral Lineage Tracing
Aug 25, 2017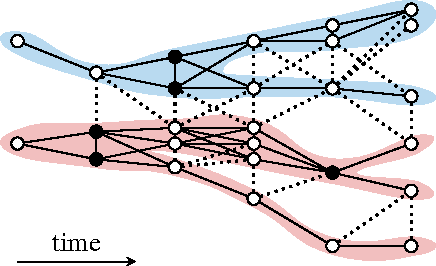
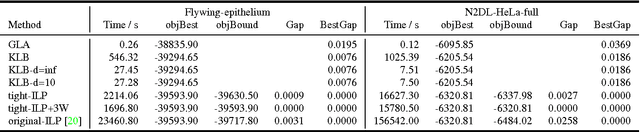
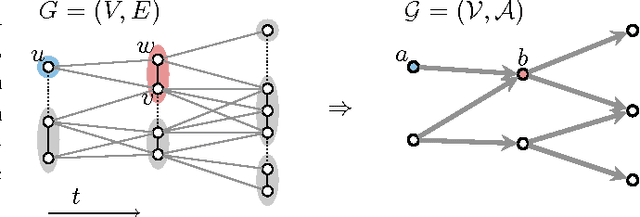
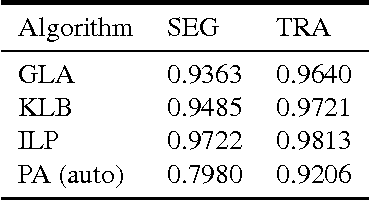
Abstract:Lineage tracing, the joint segmentation and tracking of living cells as they move and divide in a sequence of light microscopy images, is a challenging task. Jug et al. have proposed a mathematical abstraction of this task, the moral lineage tracing problem (MLTP), whose feasible solutions define both a segmentation of every image and a lineage forest of cells. Their branch-and-cut algorithm, however, is prone to many cuts and slow convergence for large instances. To address this problem, we make three contributions: (i) we devise the first efficient primal feasible local search algorithms for the MLTP, (ii) we improve the branch-and-cut algorithm by separating tighter cutting planes and by incorporating our primal algorithms, (iii) we show in experiments that our algorithms find accurate solutions on the problem instances of Jug et al. and scale to larger instances, leveraging moral lineage tracing to practical significance.
Deep-FExt: Deep Feature Extraction for Vessel Segmentation and Centerline Prediction
Apr 12, 2017
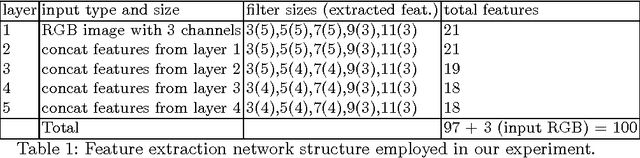
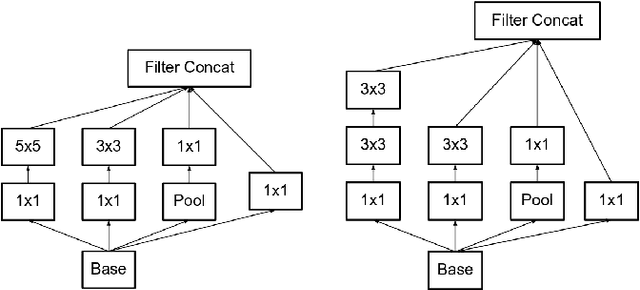
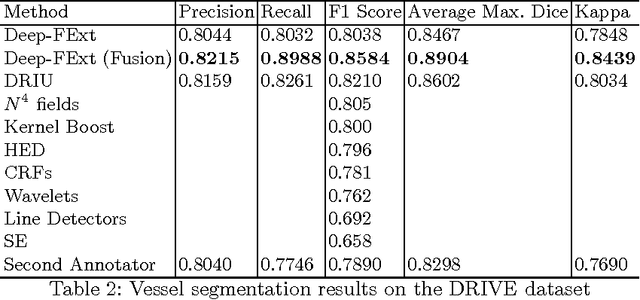
Abstract:Feature extraction is a very crucial task in image and pixel (voxel) classification and regression in biomedical image modeling. In this work we present a machine learning based feature extraction scheme based on inception models for pixel classification tasks. We extract features under multi-scale and multi-layer schemes through convolutional operators. Layers of Fully Convolutional Network are later stacked on this feature extraction layers and trained end-to-end for the purpose of classification. We test our model on the DRIVE and STARE public data sets for the purpose of segmentation and centerline detection and it out performs most existing hand crafted or deterministic feature schemes found in literature. We achieve an average maximum Dice of 0.85 on the DRIVE data set which out performs the scores from the second human annotator of this data set. We also achieve an average maximum Dice of 0.85 and kappa of 0.84 on the STARE data set. Though these datasets are mainly 2-D we also propose ways of extending this feature extraction scheme to handle 3-D datasets.
* 9 pages
Automated Unsupervised Segmentation of Liver Lesions in CT scans via Cahn-Hilliard Phase Separation
Apr 07, 2017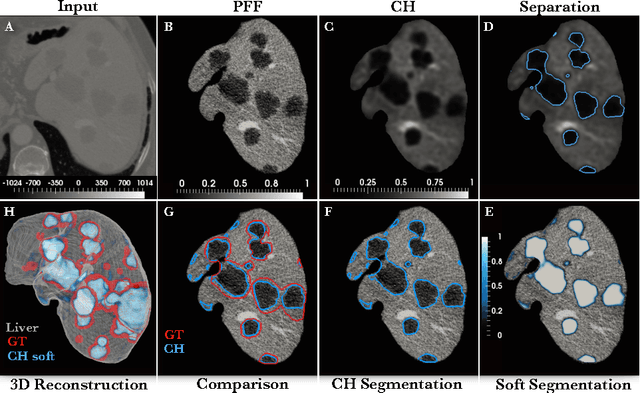
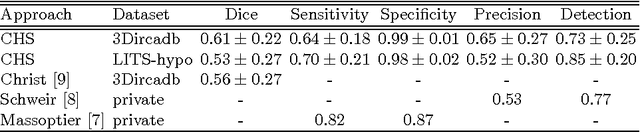
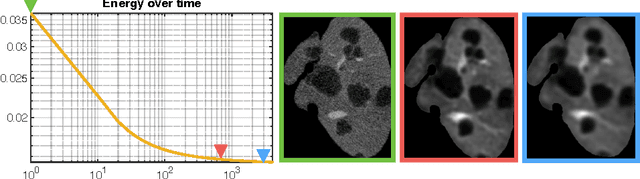
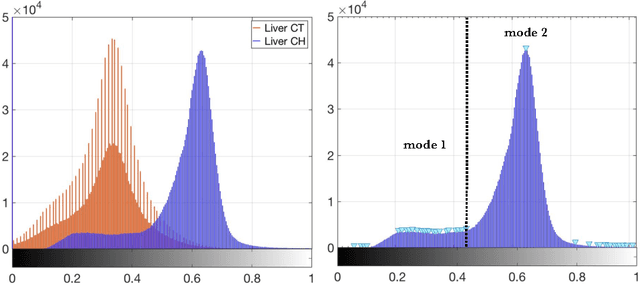
Abstract:The segmentation of liver lesions is crucial for detection, diagnosis and monitoring progression of liver cancer. However, design of accurate automated methods remains challenging due to high noise in CT scans, low contrast between liver and lesions, as well as large lesion variability. We propose a 3D automatic, unsupervised method for liver lesions segmentation using a phase separation approach. It is assumed that liver is a mixture of two phases: healthy liver and lesions, represented by different image intensities polluted by noise. The Cahn-Hilliard equation is used to remove the noise and separate the mixture into two distinct phases with well-defined interfaces. This simplifies the lesion detection and segmentation task drastically and enables to segment liver lesions by thresholding the Cahn-Hilliard solution. The method was tested on 3Dircadb and LITS dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge