Giles Tetteh
Department of Diagnostic and Interventional Neuroradiology, School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Germany, Department of Informatics, Technical University of Munich, Germany
A Deep Learning Approach to Predicting Collateral Flow in Stroke Patients Using Radiomic Features from Perfusion Images
Oct 24, 2021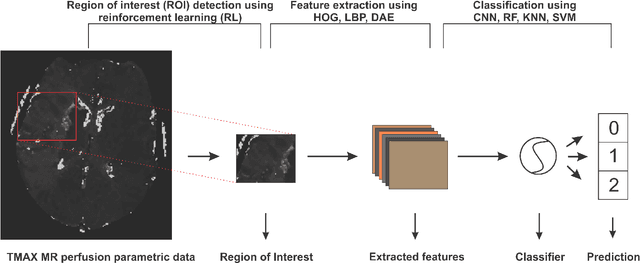
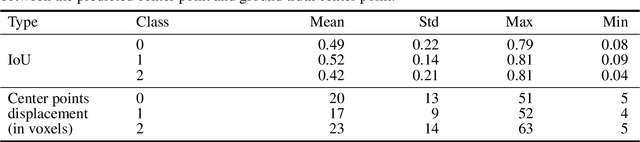
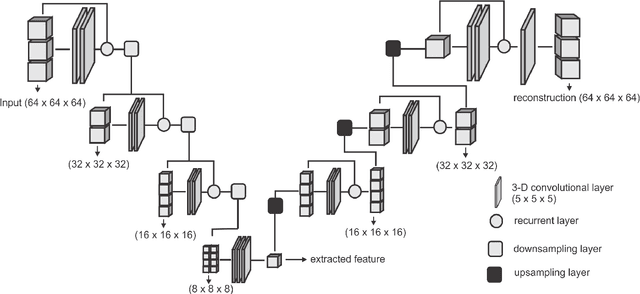
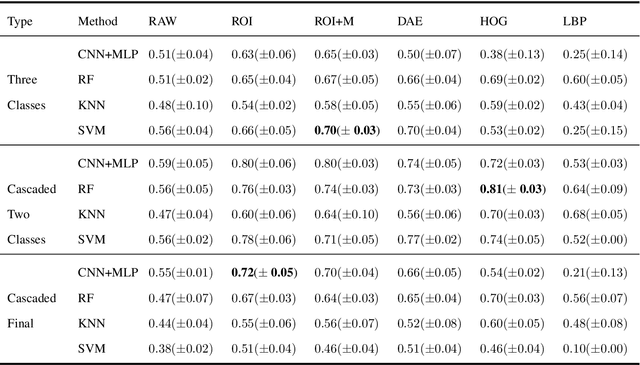
Abstract:Collateral circulation results from specialized anastomotic channels which are capable of providing oxygenated blood to regions with compromised blood flow caused by ischemic injuries. The quality of collateral circulation has been established as a key factor in determining the likelihood of a favorable clinical outcome and goes a long way to determine the choice of stroke care model - that is the decision to transport or treat eligible patients immediately. Though there exist several imaging methods and grading criteria for quantifying collateral blood flow, the actual grading is mostly done through manual inspection of the acquired images. This approach is associated with a number of challenges. First, it is time-consuming - the clinician needs to scan through several slices of images to ascertain the region of interest before deciding on what severity grade to assign to a patient. Second, there is a high tendency for bias and inconsistency in the final grade assigned to a patient depending on the experience level of the clinician. We present a deep learning approach to predicting collateral flow grading in stroke patients based on radiomic features extracted from MR perfusion data. First, we formulate a region of interest detection task as a reinforcement learning problem and train a deep learning network to automatically detect the occluded region within the 3D MR perfusion volumes. Second, we extract radiomic features from the obtained region of interest through local image descriptors and denoising auto-encoders. Finally, we apply a convolutional neural network and other machine learning classifiers to the extracted radiomic features to automatically predict the collateral flow grading of the given patient volume as one of three severity classes - no flow (0), moderate flow (1), and good flow (2)...
A Computed Tomography Vertebral Segmentation Dataset with Anatomical Variations and Multi-Vendor Scanner Data
Mar 10, 2021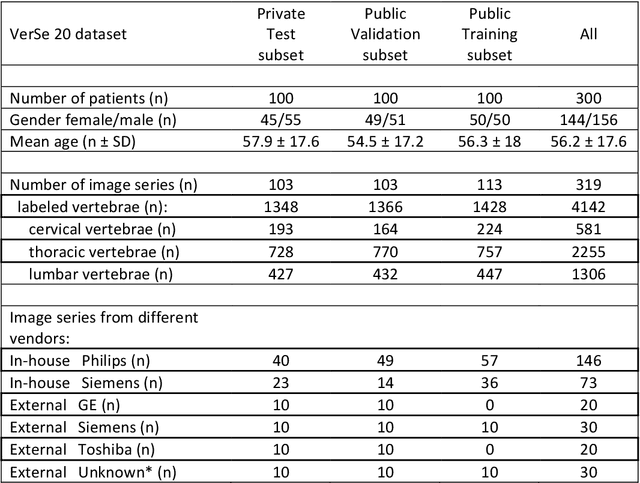
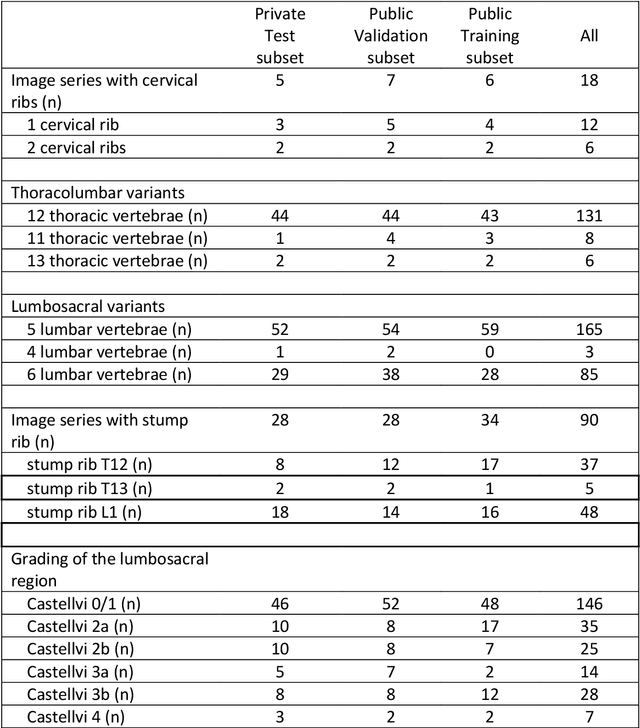
Abstract:With the advent of deep learning algorithms, fully automated radiological image analysis is within reach. In spine imaging, several atlas- and shape-based as well as deep learning segmentation algorithms have been proposed, allowing for subsequent automated analysis of morphology and pathology. The first Large Scale Vertebrae Segmentation Challenge (VerSe 2019) showed that these perform well on normal anatomy, but fail in variants not frequently present in the training dataset. Building on that experience, we report on the largely increased VerSe 2020 dataset and results from the second iteration of the VerSe challenge (MICCAI 2020, Lima, Peru). VerSe 2020 comprises annotated spine computed tomography (CT) images from 300 subjects with 4142 fully visualized and annotated vertebrae, collected across multiple centres from four different scanner manufacturers, enriched with cases that exhibit anatomical variants such as enumeration abnormalities (n=77) and transitional vertebrae (n=161). Metadata includes vertebral labelling information, voxel-level segmentation masks obtained with a human-machine hybrid algorithm and anatomical ratings, to enable the development and benchmarking of robust and accurate segmentation algorithms.
A distance-based loss for smooth and continuous skin layer segmentation in optoacoustic images
Jul 10, 2020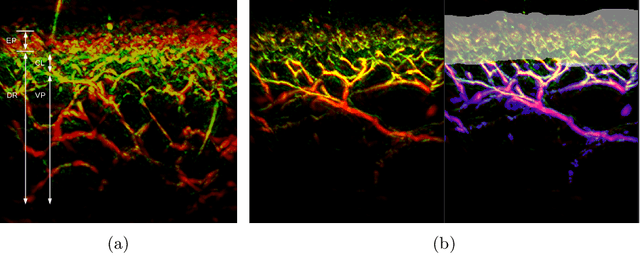
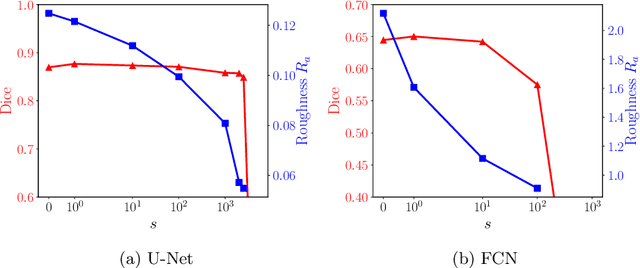

Abstract:Raster-scan optoacoustic mesoscopy (RSOM) is a powerful, non-invasive optical imaging technique for functional, anatomical, and molecular skin and tissue analysis. However, both the manual and the automated analysis of such images are challenging, because the RSOM images have very low contrast, poor signal to noise ratio, and systematic overlaps between the absorption spectra of melanin and hemoglobin. Nonetheless, the segmentation of the epidermis layer is a crucial step for many downstream medical and diagnostic tasks, such as vessel segmentation or monitoring of cancer progression. We propose a novel, shape-specific loss function that overcomes discontinuous segmentations and achieves smooth segmentation surfaces while preserving the same volumetric Dice and IoU. Further, we validate our epidermis segmentation through the sensitivity of vessel segmentation. We found a 20 $\%$ improvement in Dice for vessel segmentation tasks when the epidermis mask is provided as additional information to the vessel segmentation network.
clDice -- a Topology-Preserving Loss Function for Tubular Structure Segmentation
Mar 29, 2020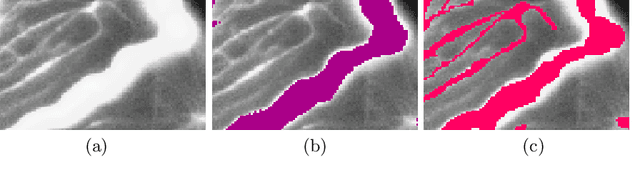
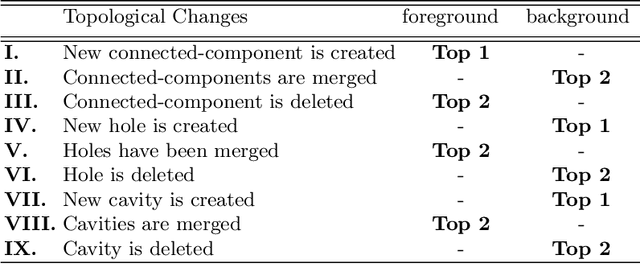
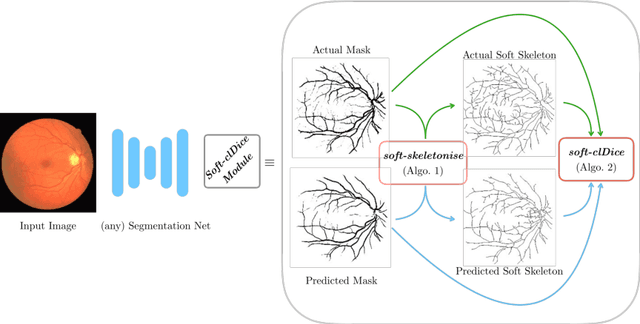
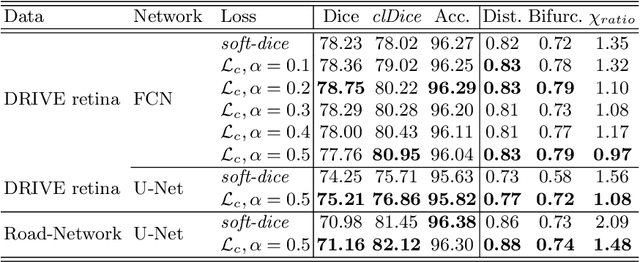
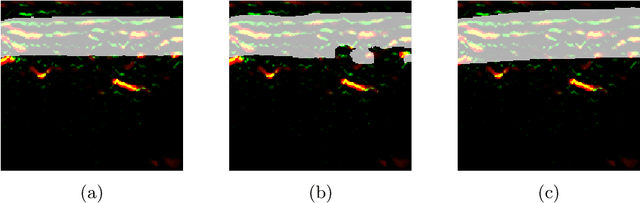
Abstract:Accurate segmentation of tubular, network-like structures, such as vessels, neurons, or roads, is relevant to many fields of research. For such structures, the topology is their most important characteristic, e.g. preserving connectedness: in case of vascular networks, missing a connected vessel entirely alters the blood-flow dynamics. We introduce a novel similarity measure termed clDice, which is calculated on the intersection of the segmentation masks and their (morphological) skeletons. Crucially, we theoretically prove that clDice guarantees topological correctness for binary 2D and 3D segmentation. Extending this, we propose a computationally efficient, differentiable soft-clDice as a loss function for training arbitrary neural segmentation networks. We benchmark the soft-clDice loss for segmentation on four public datasets (2D and 3D). Training on soft-clDice leads to segmentation with more accurate connectivity information, higher graph similarity, and better volumetric scores.
VerSe: A Vertebrae Labelling and Segmentation Benchmark
Jan 24, 2020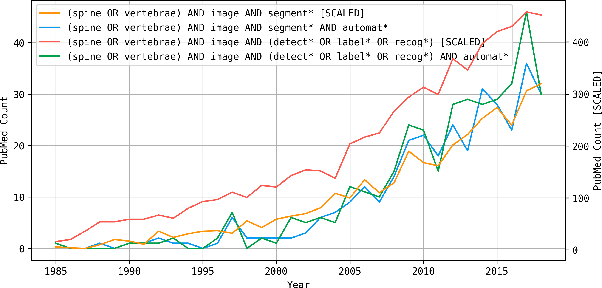
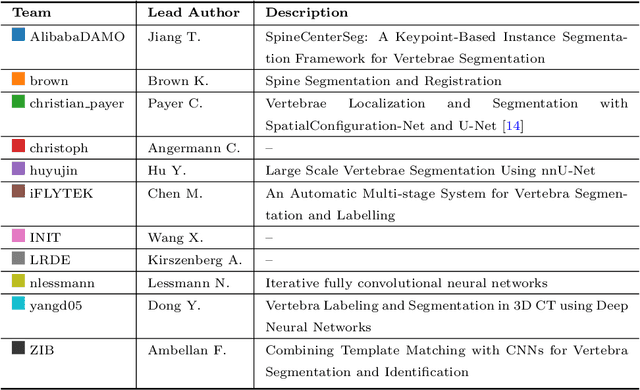
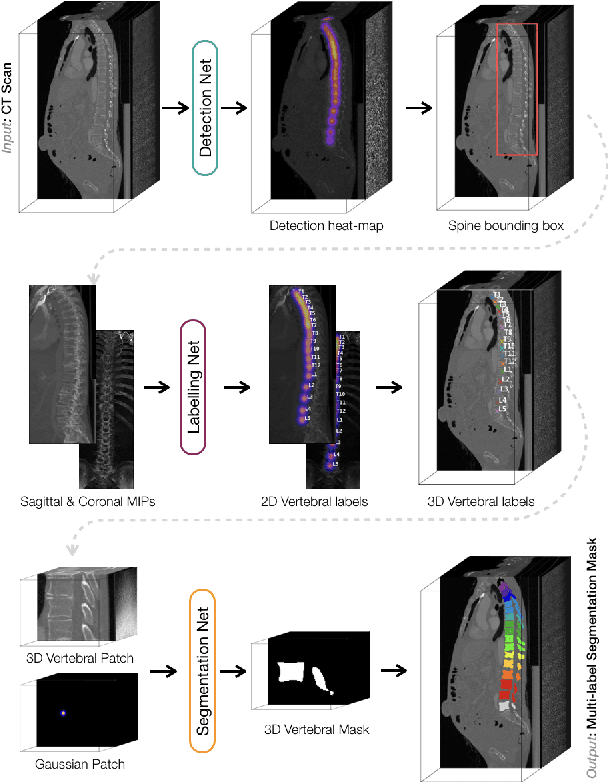
Abstract:In this paper we report the challenge set-up and results of the Large Scale Vertebrae Segmentation Challenge (VerSe) organized in conjunction with the MICCAI 2019. The challenge consisted of two tasks, vertebrae labelling and vertebrae segmentation. For this a total of 160 multidetector CT scan cohort closely resembling clinical setting was prepared and was annotated at a voxel-level by a human-machine hybrid algorithm. In this paper we also present the annotation protocol and the algorithm that aided the medical experts in the annotation process. Eleven fully automated algorithms were benchmarked on this data with the best performing algorithm achieving a vertebrae identification rate of 95% and a Dice coefficient of 90%. VerSe'19 is an open-call challenge at its image data along with the annotations and evaluation tools will continue to be publicly accessible through its online portal.
Direct Estimation of Pharmacokinetic Parameters from DCE-MRI using Deep CNN with Forward Physical Model Loss
Jun 12, 2018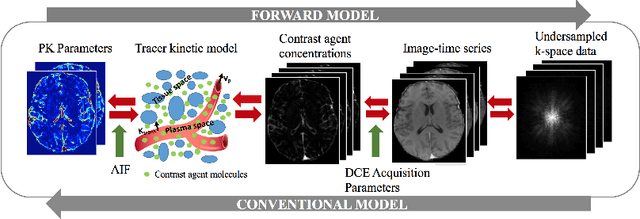
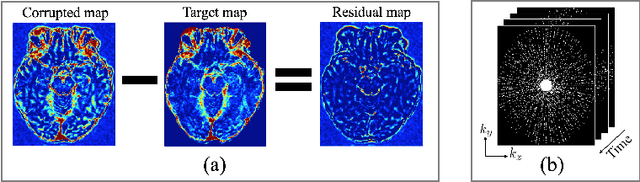
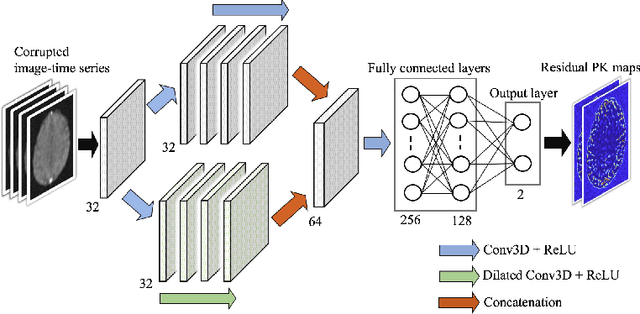
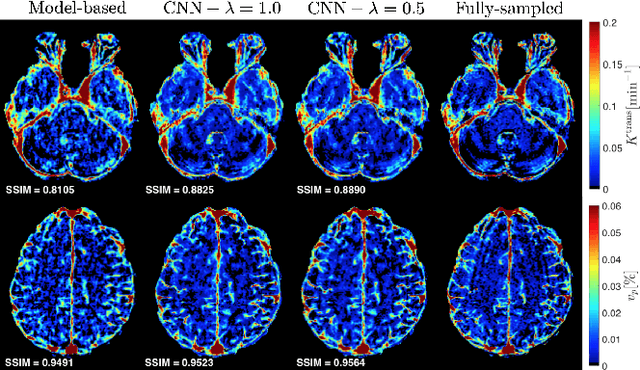
Abstract:Dynamic contrast-enhanced (DCE) MRI is an evolving imaging technique that provides a quantitative measure of pharmacokinetic (PK) parameters in body tissues, in which series of T1-weighted images are collected following the administration of a paramagnetic contrast agent. Unfortunately, in many applications, conventional clinical DCE-MRI suffers from low spatiotemporal resolution and insufficient volume coverage. In this paper, we propose a novel deep learning based approach to directly estimate the PK parameters from undersampled DCE-MRI data. Specifically, we design a custom loss function where we incorporate a forward physical model that relates the PK parameters to corrupted image-time series obtained due to subsampling in k-space. This allows the network to directly exploit the knowledge of true contrast agent kinetics in the training phase, and hence provide more accurate restoration of PK parameters. Experiments on clinical brain DCE datasets demonstrate the efficacy of our approach in terms of fidelity of PK parameter reconstruction and significantly faster parameter inference compared to a model-based iterative reconstruction method.
DeepASL: Kinetic Model Incorporated Loss for Denoising Arterial Spin Labeled MRI via Deep Residual Learning
Jun 12, 2018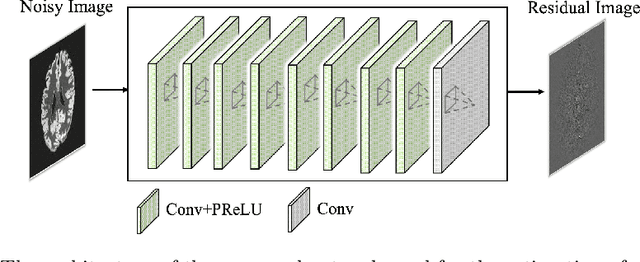
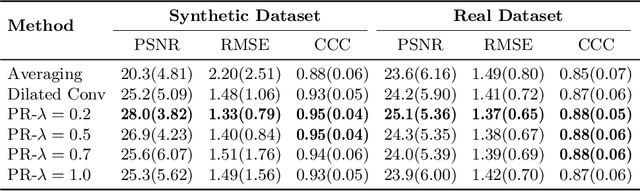
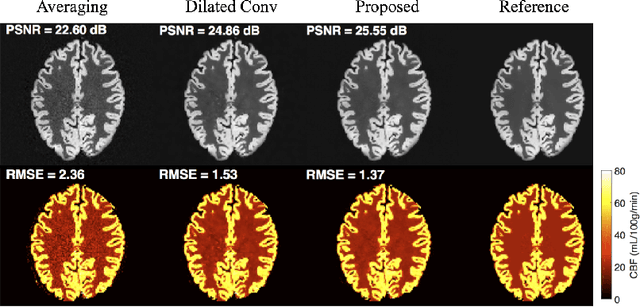
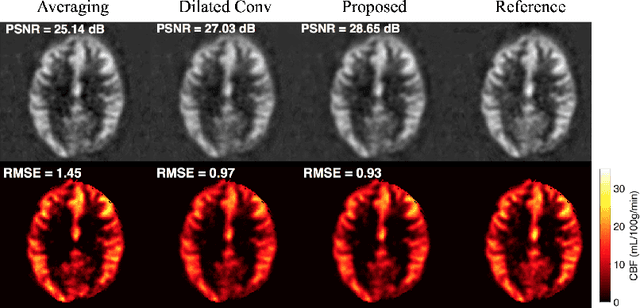
Abstract:Arterial spin labeling (ASL) allows to quantify the cerebral blood flow (CBF) by magnetic labeling of the arterial blood water. ASL is increasingly used in clinical studies due to its noninvasiveness, repeatability and benefits in quantification. However, ASL suffers from an inherently low-signal-to-noise ratio (SNR) requiring repeated measurements of control/spin-labeled (C/L) pairs to achieve a reasonable image quality, which in return increases motion sensitivity. This leads to clinically prolonged scanning times increasing the risk of motion artifacts. Thus, there is an immense need of advanced imaging and processing techniques in ASL. In this paper, we propose a novel deep learning based approach to improve the perfusion-weighted image quality obtained from a subset of all available pairwise C/L subtractions. Specifically, we train a deep fully convolutional network (FCN) to learn a mapping from noisy perfusion-weighted image and its subtraction (residual) from the clean image. Additionally, we incorporate the CBF estimation model in the loss function during training, which enables the network to produce high quality images while simultaneously enforcing the CBF estimates to be as close as reference CBF values. Extensive experiments on synthetic and clinical ASL datasets demonstrate the effectiveness of our method in terms of improved ASL image quality, accurate CBF parameter estimation and considerably small computation time during testing.
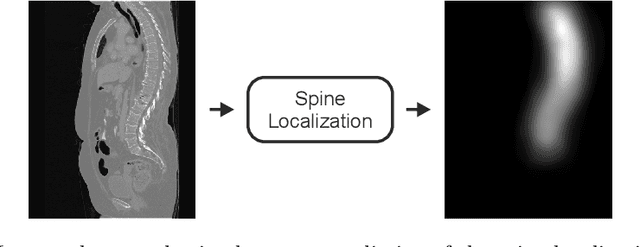
Btrfly Net: Vertebrae Labelling with Energy-based Adversarial Learning of Local Spine Prior
Apr 04, 2018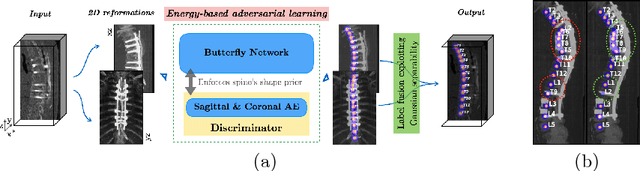
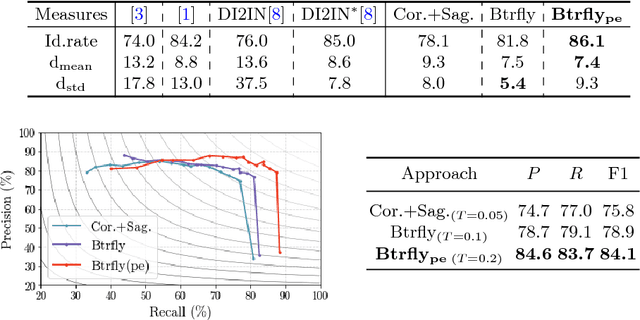
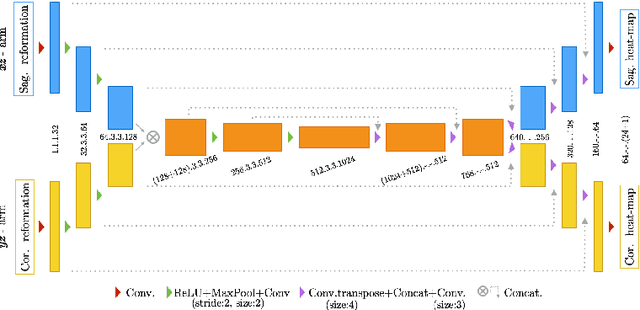
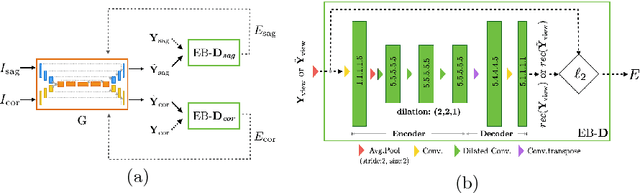
Abstract:Robust localisation and identification of vertebrae is an essential part of automated spine analysis. The contribution of this work to the task is two-fold: (1) Inspired by the human expert, we hypothesise that a sagittal and coronal reformation of the spine contain sufficient information for labelling the vertebrae. Thereby, we propose a butterfly-shaped network architecture (termed Btrfly Net) that efficiently combines the information across the reformations. (2) Underpinning the Btrfly net, we present an energy-based adversarial training regime that encodes the local spine structure as an anatomical prior into the network, thereby enabling it to achieve state-of-art performance in all standard metrics on a benchmark dataset of 302 scans without any post-processing during inference.
DeepVesselNet: Vessel Segmentation, Centerline Prediction, and Bifurcation Detection in 3-D Angiographic Volumes
Mar 25, 2018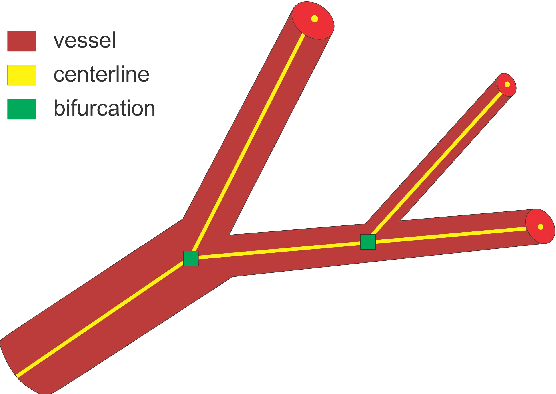
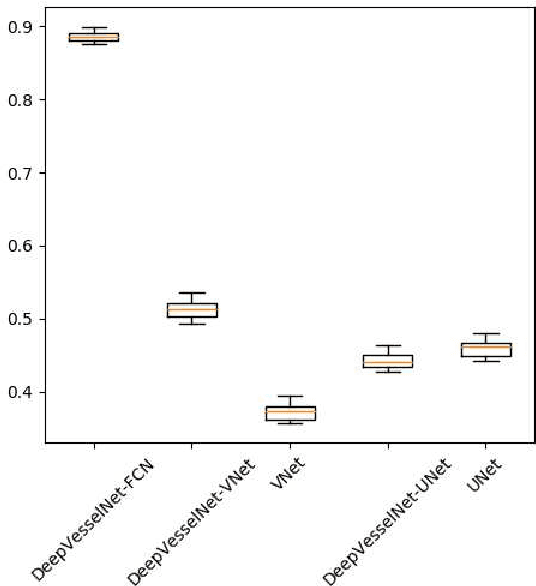
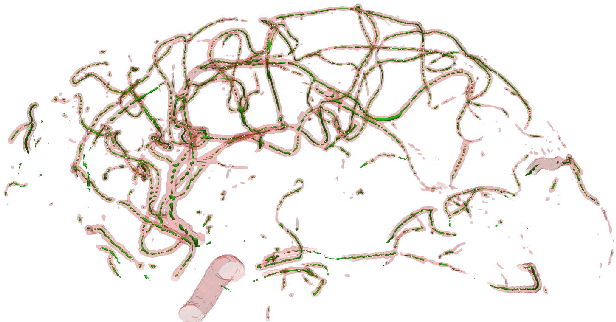
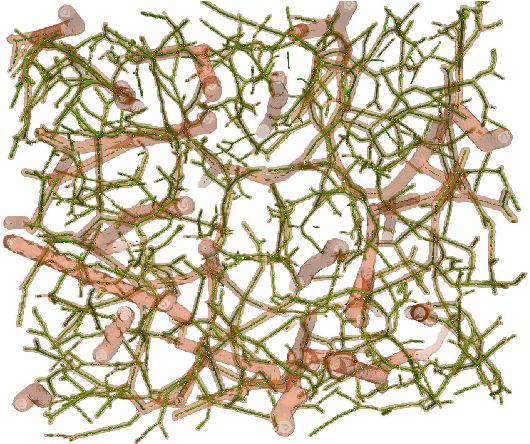
Abstract:We present DeepVesselNet, an architecture tailored to the challenges to be addressed when extracting vessel networks and corresponding features in 3-D angiography using deep learning. We discuss the problems of low execution speed and high memory requirements associated with full 3-D convolutional networks, high class imbalance arising from low percentage (less than 3%) of vessel voxels, and unavailability of accurately annotated training data - and offer solutions that are the building blocks of DeepVesselNet. First, we formulate 2-D orthogonal cross-hair filters which make use of 3-D context information. Second, we introduce a class balancing cross-entropy score with false positive rate correction to handle the high class imbalance and high false positive rate problems associated with existing loss functions. Finally, we generate synthetic dataset using a computational angiogenesis model, capable of generating vascular networks under physiological constraints on local network structure and topology, and use these data for transfer learning. DeepVesselNet is optimized for segmenting vessels, predicting centerlines, and localizing bifurcations. We test the performance on a range of angiographic volumes including clinical Time-of-Flight MRA data of the human brain, as well as synchrotron radiation X-ray tomographic microscopy scans of the rat brain. Our experiments show that, by replacing 3-D filters with 2-D orthogonal cross-hair filters in our network, speed is improved by 23% while accuracy is maintained. Our class balancing metric is crucial for training the network and pre-training with synthetic data helps in early convergence of the training process.
Deep-FExt: Deep Feature Extraction for Vessel Segmentation and Centerline Prediction
Apr 12, 2017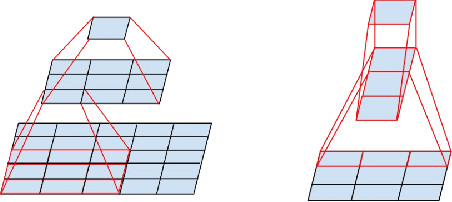
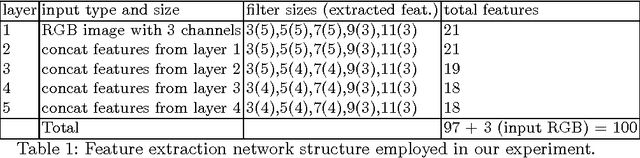
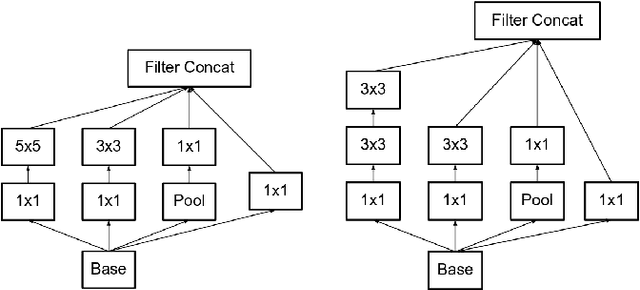
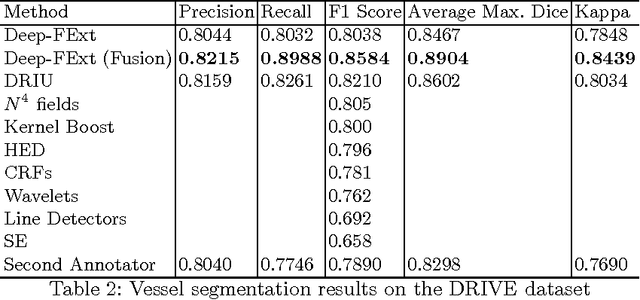
Abstract:Feature extraction is a very crucial task in image and pixel (voxel) classification and regression in biomedical image modeling. In this work we present a machine learning based feature extraction scheme based on inception models for pixel classification tasks. We extract features under multi-scale and multi-layer schemes through convolutional operators. Layers of Fully Convolutional Network are later stacked on this feature extraction layers and trained end-to-end for the purpose of classification. We test our model on the DRIVE and STARE public data sets for the purpose of segmentation and centerline detection and it out performs most existing hand crafted or deterministic feature schemes found in literature. We achieve an average maximum Dice of 0.85 on the DRIVE data set which out performs the scores from the second human annotator of this data set. We also achieve an average maximum Dice of 0.85 and kappa of 0.84 on the STARE data set. Though these datasets are mainly 2-D we also propose ways of extending this feature extraction scheme to handle 3-D datasets.
* 9 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge