Kai Ma
School of Electrical Engineering, Yanshan University, Qinhuangdao, China
All-Around Real Label Supervision: Cyclic Prototype Consistency Learning for Semi-supervised Medical Image Segmentation
Sep 28, 2021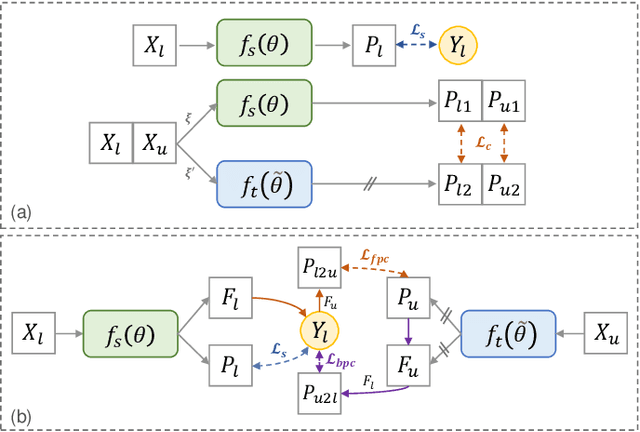
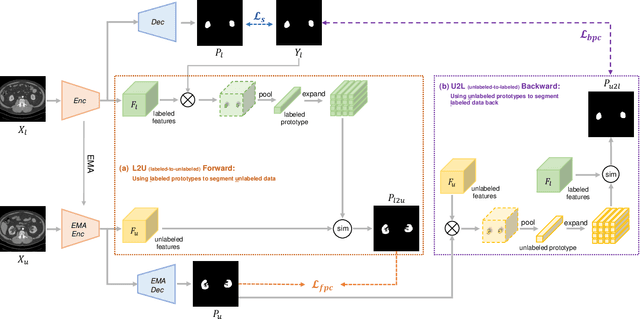
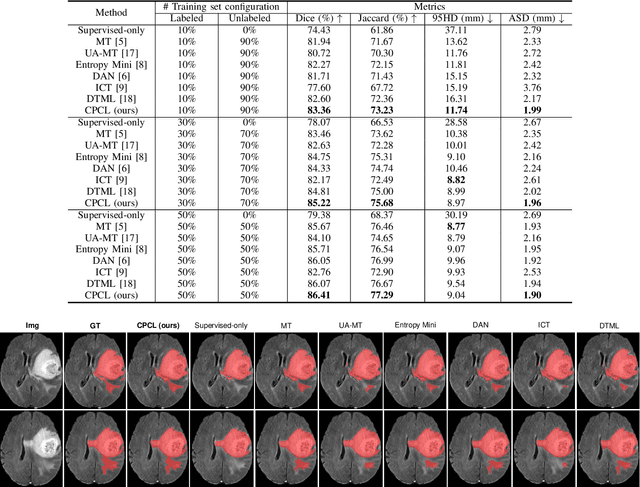
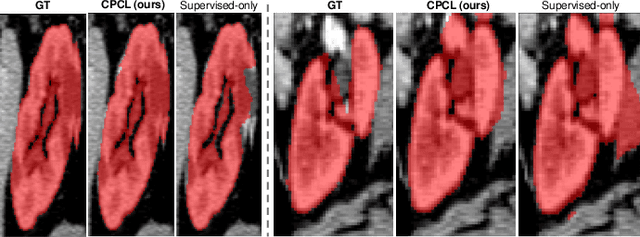
Abstract:Semi-supervised learning has substantially advanced medical image segmentation since it alleviates the heavy burden of acquiring the costly expert-examined annotations. Especially, the consistency-based approaches have attracted more attention for their superior performance, wherein the real labels are only utilized to supervise their paired images via supervised loss while the unlabeled images are exploited by enforcing the perturbation-based \textit{"unsupervised"} consistency without explicit guidance from those real labels. However, intuitively, the expert-examined real labels contain more reliable supervision signals. Observing this, we ask an unexplored but interesting question: can we exploit the unlabeled data via explicit real label supervision for semi-supervised training? To this end, we discard the previous perturbation-based consistency but absorb the essence of non-parametric prototype learning. Based on the prototypical network, we then propose a novel cyclic prototype consistency learning (CPCL) framework, which is constructed by a labeled-to-unlabeled (L2U) prototypical forward process and an unlabeled-to-labeled (U2L) backward process. Such two processes synergistically enhance the segmentation network by encouraging more discriminative and compact features. In this way, our framework turns previous \textit{"unsupervised"} consistency into new \textit{"supervised"} consistency, obtaining the \textit{"all-around real label supervision"} property of our method. Extensive experiments on brain tumor segmentation from MRI and kidney segmentation from CT images show that our CPCL can effectively exploit the unlabeled data and outperform other state-of-the-art semi-supervised medical image segmentation methods.
Training Automatic View Planner for Cardiac MR Imaging via Self-Supervision by Spatial Relationship between Views
Sep 24, 2021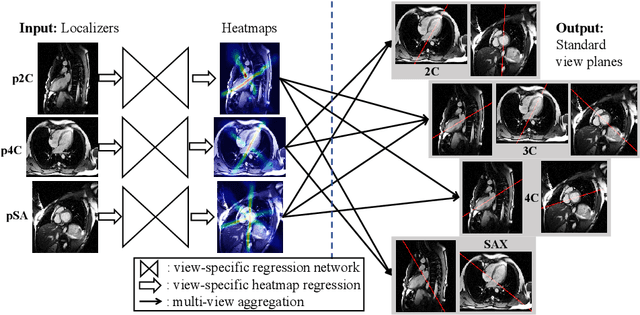
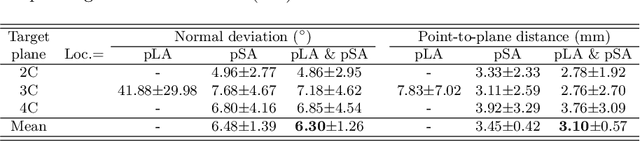
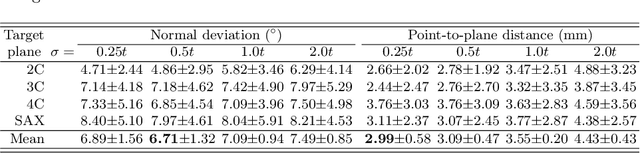
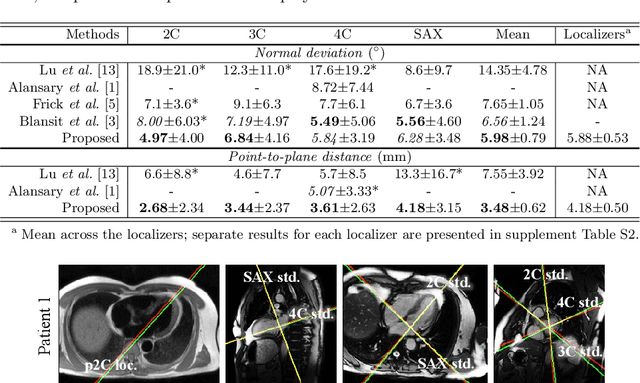
Abstract:View planning for the acquisition of cardiac magnetic resonance imaging (CMR) requires acquaintance with the cardiac anatomy and remains a challenging task in clinical practice. Existing approaches to its automation relied either on an additional volumetric image not typically acquired in clinic routine, or on laborious manual annotations of cardiac structural landmarks. This work presents a clinic-compatible and annotation-free system for automatic CMR view planning. The system mines the spatial relationship -- more specifically, locates and exploits the intersecting lines -- between the source and target views, and trains deep networks to regress heatmaps defined by these intersecting lines. As the spatial relationship is self-contained in properly stored data, e.g., in the DICOM format, the need for manual annotation is eliminated. Then, a multi-view planning strategy is proposed to aggregate information from the predicted heatmaps for all the source views of a target view, for a globally optimal prescription. The multi-view aggregation mimics the similar strategy practiced by skilled human prescribers. Experimental results on 181 clinical CMR exams show that our system achieves superior accuracy to existing approaches including conventional atlas-based and newer deep learning based ones, in prescribing four standard CMR views. The mean angle difference and point-to-plane distance evaluated against the ground truth planes are 5.98 degrees and 3.48 mm, respectively.
InDuDoNet: An Interpretable Dual Domain Network for CT Metal Artifact Reduction
Sep 11, 2021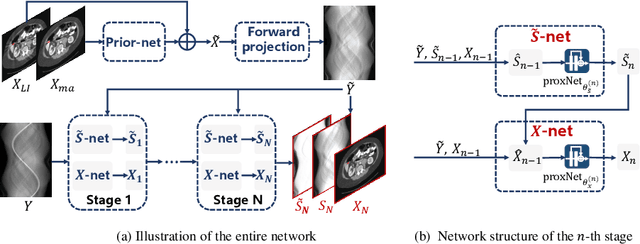
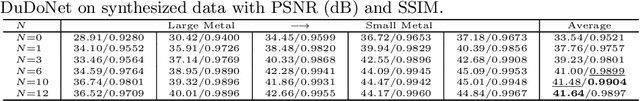
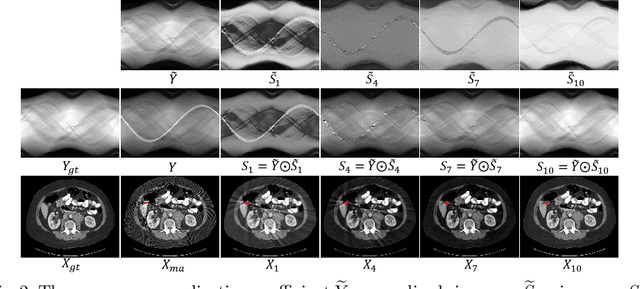
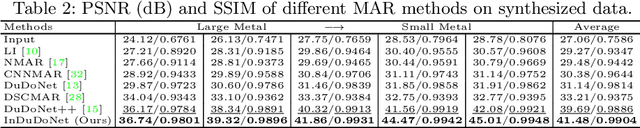
Abstract:For the task of metal artifact reduction (MAR), although deep learning (DL)-based methods have achieved promising performances, most of them suffer from two problems: 1) the CT imaging geometry constraint is not fully embedded into the network during training, leaving room for further performance improvement; 2) the model interpretability is lack of sufficient consideration. Against these issues, we propose a novel interpretable dual domain network, termed as InDuDoNet, which combines the advantages of model-driven and data-driven methodologies. Specifically, we build a joint spatial and Radon domain reconstruction model and utilize the proximal gradient technique to design an iterative algorithm for solving it. The optimization algorithm only consists of simple computational operators, which facilitate us to correspondingly unfold iterative steps into network modules and thus improve the interpretablility of the framework. Extensive experiments on synthesized and clinical data show the superiority of our InDuDoNet. Code is available in \url{https://github.com/hongwang01/InDuDoNet}.%method on the tasks of MAR and downstream multi-class pelvic fracture segmentation.
Multi-Anchor Active Domain Adaptation for Semantic Segmentation
Aug 18, 2021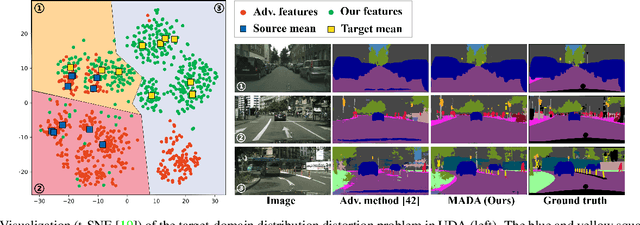
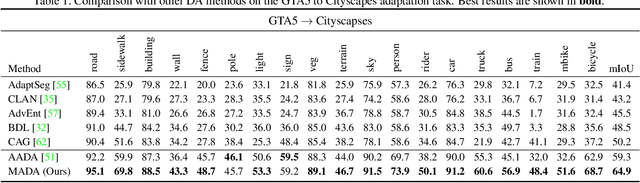
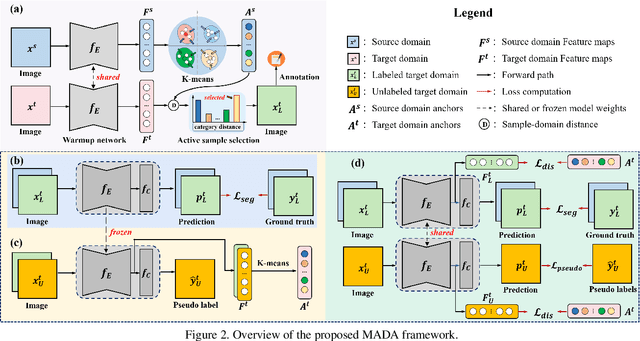
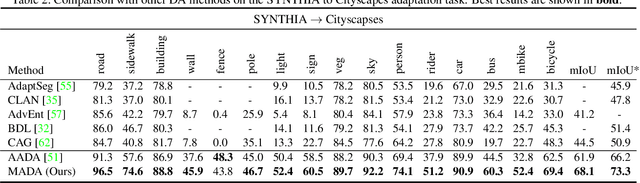
Abstract:Unsupervised domain adaption has proven to be an effective approach for alleviating the intensive workload of manual annotation by aligning the synthetic source-domain data and the real-world target-domain samples. Unfortunately, mapping the target-domain distribution to the source-domain unconditionally may distort the essential structural information of the target-domain data. To this end, we firstly propose to introduce a novel multi-anchor based active learning strategy to assist domain adaptation regarding the semantic segmentation task. By innovatively adopting multiple anchors instead of a single centroid, the source domain can be better characterized as a multimodal distribution, thus more representative and complimentary samples are selected from the target domain. With little workload to manually annotate these active samples, the distortion of the target-domain distribution can be effectively alleviated, resulting in a large performance gain. The multi-anchor strategy is additionally employed to model the target-distribution. By regularizing the latent representation of the target samples compact around multiple anchors through a novel soft alignment loss, more precise segmentation can be achieved. Extensive experiments are conducted on public datasets to demonstrate that the proposed approach outperforms state-of-the-art methods significantly, along with thorough ablation study to verify the effectiveness of each component.
A New Bidirectional Unsupervised Domain Adaptation Segmentation Framework
Aug 18, 2021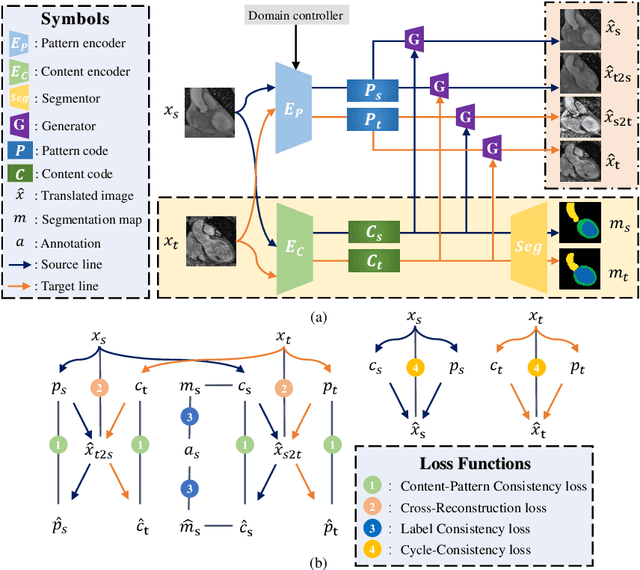
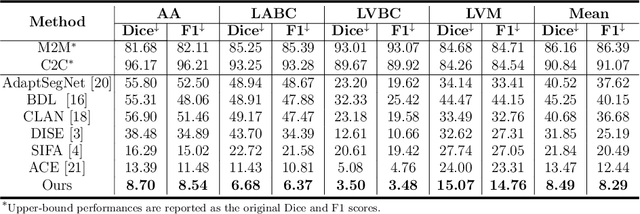
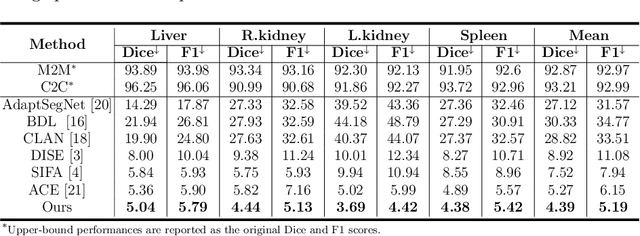
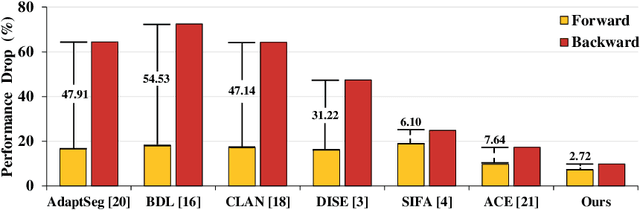
Abstract:Domain shift happens in cross-domain scenarios commonly because of the wide gaps between different domains: when applying a deep learning model well-trained in one domain to another target domain, the model usually performs poorly. To tackle this problem, unsupervised domain adaptation (UDA) techniques are proposed to bridge the gap between different domains, for the purpose of improving model performance without annotation in the target domain. Particularly, UDA has a great value for multimodal medical image analysis, where annotation difficulty is a practical concern. However, most existing UDA methods can only achieve satisfactory improvements in one adaptation direction (e.g., MRI to CT), but often perform poorly in the other (CT to MRI), limiting their practical usage. In this paper, we propose a bidirectional UDA (BiUDA) framework based on disentangled representation learning for equally competent two-way UDA performances. This framework employs a unified domain-aware pattern encoder which not only can adaptively encode images in different domains through a domain controller, but also improve model efficiency by eliminating redundant parameters. Furthermore, to avoid distortion of contents and patterns of input images during the adaptation process, a content-pattern consistency loss is introduced. Additionally, for better UDA segmentation performance, a label consistency strategy is proposed to provide extra supervision by recomposing target-domain-styled images and corresponding source-domain annotations. Comparison experiments and ablation studies conducted on two public datasets demonstrate the superiority of our BiUDA framework to current state-of-the-art UDA methods and the effectiveness of its novel designs. By successfully addressing two-way adaptations, our BiUDA framework offers a flexible solution of UDA techniques to the real-world scenario.
RECIST-Net: Lesion detection via grouping keypoints on RECIST-based annotation
Jul 19, 2021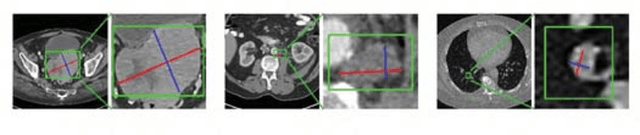
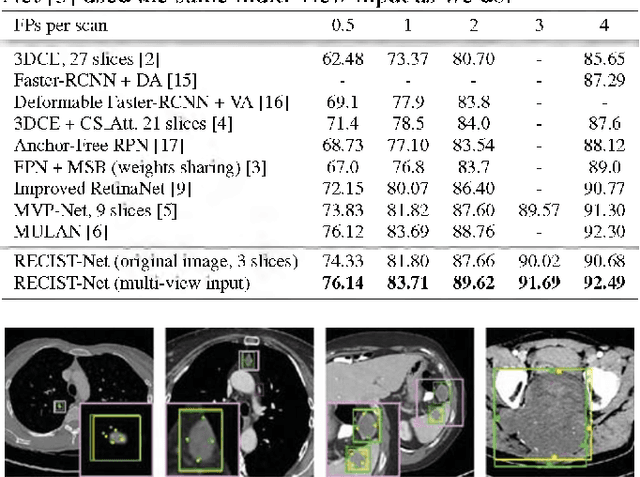
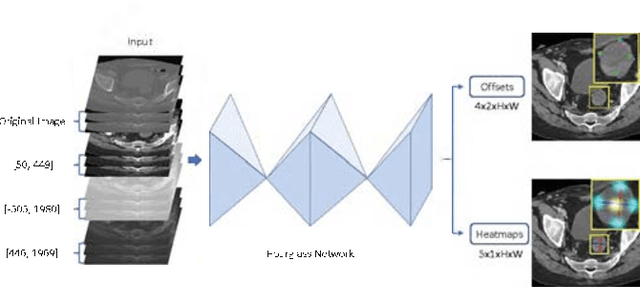
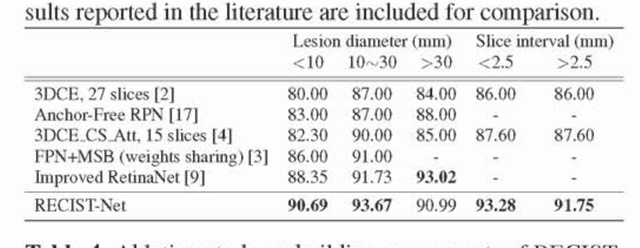
Abstract:Universal lesion detection in computed tomography (CT) images is an important yet challenging task due to the large variations in lesion type, size, shape, and appearance. Considering that data in clinical routine (such as the DeepLesion dataset) are usually annotated with a long and a short diameter according to the standard of Response Evaluation Criteria in Solid Tumors (RECIST) diameters, we propose RECIST-Net, a new approach to lesion detection in which the four extreme points and center point of the RECIST diameters are detected. By detecting a lesion as keypoints, we provide a more conceptually straightforward formulation for detection, and overcome several drawbacks (e.g., requiring extensive effort in designing data-appropriate anchors and losing shape information) of existing bounding-box-based methods while exploring a single-task, one-stage approach compared to other RECIST-based approaches. Experiments show that RECIST-Net achieves a sensitivity of 92.49% at four false positives per image, outperforming other recent methods including those using multi-task learning.
Double-Uncertainty Assisted Spatial and Temporal Regularization Weighting for Learning-based Registration
Jul 15, 2021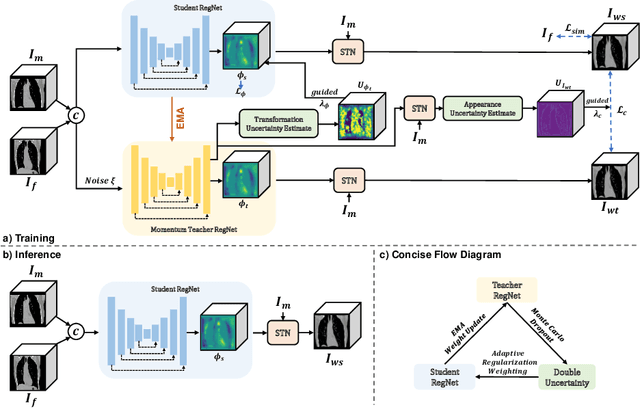
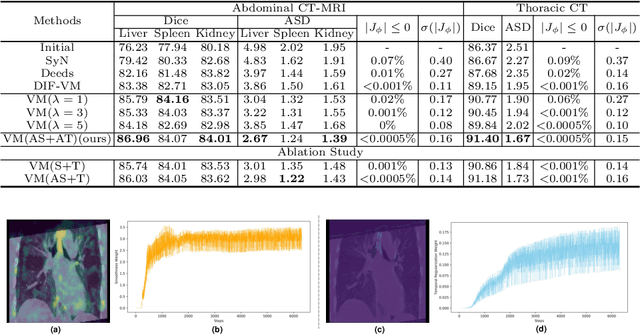
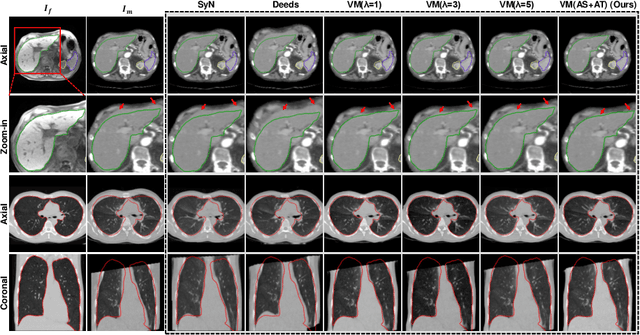
Abstract:In order to tackle the difficulty associated with the ill-posed nature of the image registration problem, researchers use regularization to constrain the solution space. For most learning-based registration approaches, the regularization usually has a fixed weight and only constrains the spatial transformation. Such convention has two limitations: (1) The regularization strength of a specific image pair should be associated with the content of the images, thus the ``one value fits all'' scheme is not ideal; (2) Only spatially regularizing the transformation (but overlooking the temporal consistency of different estimations) may not be the best strategy to cope with the ill-posedness. In this study, we propose a mean-teacher based registration framework. This framework incorporates an additional \textit{temporal regularization} term by encouraging the teacher model's temporal ensemble prediction to be consistent with that of the student model. At each training step, it also automatically adjusts the weights of the \textit{spatial regularization} and the \textit{temporal regularization} by taking account of the transformation uncertainty and appearance uncertainty derived from the perturbed teacher model. We perform experiments on multi- and uni-modal registration tasks, and the results show that our strategy outperforms the traditional and learning-based benchmark methods.
Mutual-GAN: Towards Unsupervised Cross-Weather Adaptation with Mutual Information Constraint
Jun 30, 2021
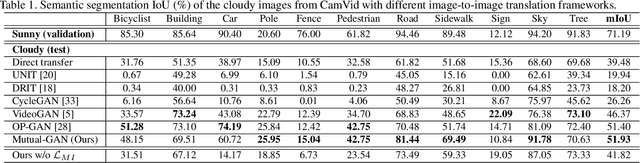
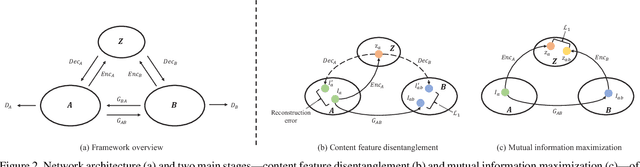
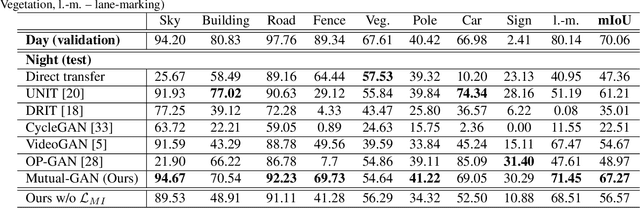
Abstract:Convolutional neural network (CNN) have proven its success for semantic segmentation, which is a core task of emerging industrial applications such as autonomous driving. However, most progress in semantic segmentation of urban scenes is reported on standard scenarios, i.e., daytime scenes with favorable illumination conditions. In practical applications, the outdoor weather and illumination are changeable, e.g., cloudy and nighttime, which results in a significant drop of semantic segmentation accuracy of CNN only trained with daytime data. In this paper, we propose a novel generative adversarial network (namely Mutual-GAN) to alleviate the accuracy decline when daytime-trained neural network is applied to videos captured under adverse weather conditions. The proposed Mutual-GAN adopts mutual information constraint to preserve image-objects during cross-weather adaptation, which is an unsolved problem for most unsupervised image-to-image translation approaches (e.g., CycleGAN). The proposed Mutual-GAN is evaluated on two publicly available driving video datasets (i.e., CamVid and SYNTHIA). The experimental results demonstrate that our Mutual-GAN can yield visually plausible translated images and significantly improve the semantic segmentation accuracy of daytime-trained deep learning network while processing videos under challenging weathers.
Residual Moment Loss for Medical Image Segmentation
Jun 27, 2021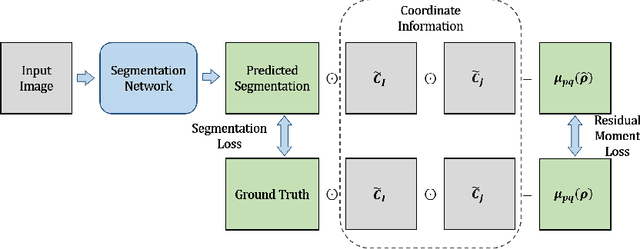
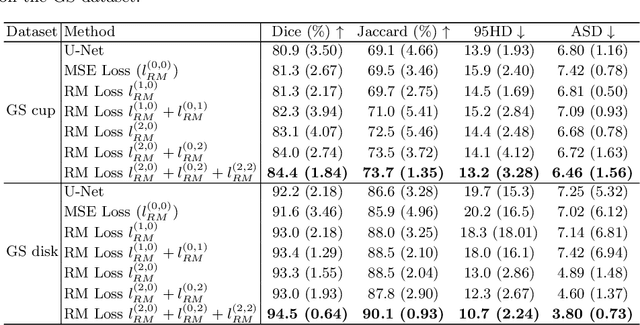
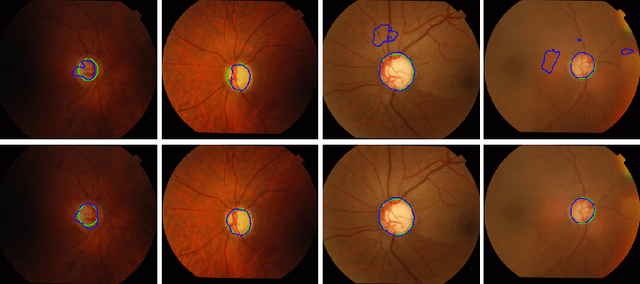
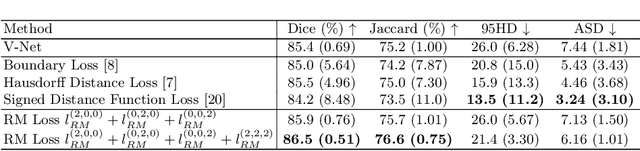
Abstract:Location information is proven to benefit the deep learning models on capturing the manifold structure of target objects, and accordingly boosts the accuracy of medical image segmentation. However, most existing methods encode the location information in an implicit way, e.g. the distance transform maps, which describe the relative distance from each pixel to the contour boundary, for the network to learn. These implicit approaches do not fully exploit the position information (i.e. absolute location) of targets. In this paper, we propose a novel loss function, namely residual moment (RM) loss, to explicitly embed the location information of segmentation targets during the training of deep learning networks. Particularly, motivated by image moments, the segmentation prediction map and ground-truth map are weighted by coordinate information. Then our RM loss encourages the networks to maintain the consistency between the two weighted maps, which promotes the segmentation networks to easily locate the targets and extract manifold-structure-related features. We validate the proposed RM loss by conducting extensive experiments on two publicly available datasets, i.e., 2D optic cup and disk segmentation and 3D left atrial segmentation. The experimental results demonstrate the effectiveness of our RM loss, which significantly boosts the accuracy of segmentation networks.
LE-NAS: Learning-based Ensenble with NAS for Dose Prediction
Jun 12, 2021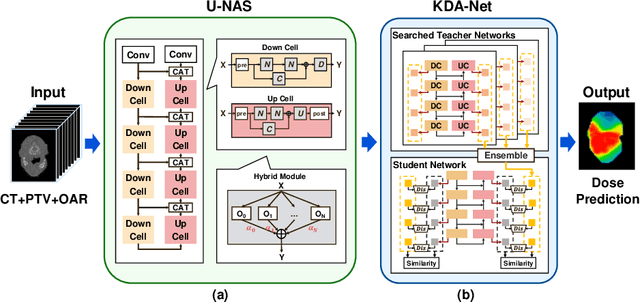
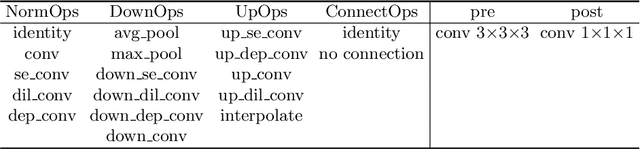
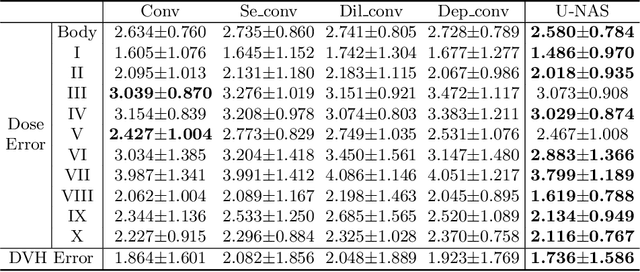
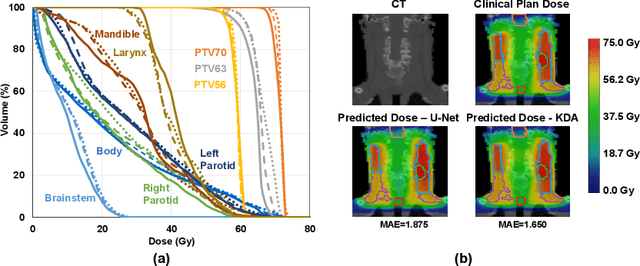
Abstract:Radiation therapy treatment planning is a complex process, as the target dose prescription and normal tissue sparing are conflicting objectives. Automated and accurate dose prediction for radiation therapy planning is in high demand. In this study, we propose a novel learning-based ensemble approach, named LE-NAS, which integrates neural architecture search (NAS) with knowledge distillation for 3D radiotherapy dose prediction. Specifically, the prediction network first exhaustively searches each block from enormous architecture space. Then, multiple architectures are selected with promising performance and diversity. To reduce the inference time, we adopt the teacher-student paradigm by treating the combination of diverse outputs from multiple searched networks as supervisions to guide the student network training. In addition, we apply adversarial learning to optimize the student network to recover the knowledge in teacher networks. To the best of our knowledge, we are the first to investigate the combination of NAS and knowledge distillation. The proposed method has been evaluated on the public OpenKBP dataset, and experimental results demonstrate the effectiveness of our method and its superior performance to the state-of-the-art method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge