Jianhua Ma
DuMeta++: Spatiotemporal Dual Meta-Learning for Generalizable Few-Shot Brain Tissue Segmentation Across Diverse Ages
Feb 06, 2026Abstract:Accurate segmentation of brain tissues from MRI scans is critical for neuroscience and clinical applications, but achieving consistent performance across the human lifespan remains challenging due to dynamic, age-related changes in brain appearance and morphology. While prior work has sought to mitigate these shifts by using self-supervised regularization with paired longitudinal data, such data are often unavailable in practice. To address this, we propose \emph{DuMeta++}, a dual meta-learning framework that operates without paired longitudinal data. Our approach integrates: (1) meta-feature learning to extract age-agnostic semantic representations of spatiotemporally evolving brain structures, and (2) meta-initialization learning to enable data-efficient adaptation of the segmentation model. Furthermore, we propose a memory-bank-based class-aware regularization strategy to enforce longitudinal consistency without explicit longitudinal supervision. We theoretically prove the convergence of our DuMeta++, ensuring stability. Experiments on diverse datasets (iSeg-2019, IBIS, OASIS, ADNI) under few-shot settings demonstrate that DuMeta++ outperforms existing methods in cross-age generalization. Code will be available at https://github.com/ladderlab-xjtu/DuMeta++.
UniSino: Physics-Driven Foundational Model for Universal CT Sinogram Standardization
Aug 25, 2025



Abstract:During raw-data acquisition in CT imaging, diverse factors can degrade the collected sinograms, with undersampling and noise leading to severe artifacts and noise in reconstructed images and compromising diagnostic accuracy. Conventional correction methods rely on manually designed algorithms or fixed empirical parameters, but these approaches often lack generalizability across heterogeneous artifact types. To address these limitations, we propose UniSino, a foundation model for universal CT sinogram standardization. Unlike existing foundational models that operate in image domain, UniSino directly standardizes data in the projection domain, which enables stronger generalization across diverse undersampling scenarios. Its training framework incorporates the physical characteristics of sinograms, enhancing generalization and enabling robust performance across multiple subtasks spanning four benchmark datasets. Experimental results demonstrate thatUniSino achieves superior reconstruction quality both single and mixed undersampling case, demonstrating exceptional robustness and generalization in sinogram enhancement for CT imaging. The code is available at: https://github.com/yqx7150/UniSino.
FEAT: A Multi-Agent Forensic AI System with Domain-Adapted Large Language Model for Automated Cause-of-Death Analysis
Aug 11, 2025Abstract:Forensic cause-of-death determination faces systemic challenges, including workforce shortages and diagnostic variability, particularly in high-volume systems like China's medicolegal infrastructure. We introduce FEAT (ForEnsic AgenT), a multi-agent AI framework that automates and standardizes death investigations through a domain-adapted large language model. FEAT's application-oriented architecture integrates: (i) a central Planner for task decomposition, (ii) specialized Local Solvers for evidence analysis, (iii) a Memory & Reflection module for iterative refinement, and (iv) a Global Solver for conclusion synthesis. The system employs tool-augmented reasoning, hierarchical retrieval-augmented generation, forensic-tuned LLMs, and human-in-the-loop feedback to ensure legal and medical validity. In evaluations across diverse Chinese case cohorts, FEAT outperformed state-of-the-art AI systems in both long-form autopsy analyses and concise cause-of-death conclusions. It demonstrated robust generalization across six geographic regions and achieved high expert concordance in blinded validations. Senior pathologists validated FEAT's outputs as comparable to those of human experts, with improved detection of subtle evidentiary nuances. To our knowledge, FEAT is the first LLM-based AI agent system dedicated to forensic medicine, offering scalable, consistent death certification while maintaining expert-level rigor. By integrating AI efficiency with human oversight, this work could advance equitable access to reliable medicolegal services while addressing critical capacity constraints in forensic systems.
Continuous Filtered Backprojection by Learnable Interpolation Network
May 03, 2025Abstract:Accurate reconstruction of computed tomography (CT) images is crucial in medical imaging field. However, there are unavoidable interpolation errors in the backprojection step of the conventional reconstruction methods, i.e., filtered-back-projection based methods, which are detrimental to the accurate reconstruction. In this study, to address this issue, we propose a novel deep learning model, named Leanable-Interpolation-based FBP or LInFBP shortly, to enhance the reconstructed CT image quality, which achieves learnable interpolation in the backprojection step of filtered backprojection (FBP) and alleviates the interpolation errors. Specifically, in the proposed LInFBP, we formulate every local piece of the latent continuous function of discrete sinogram data as a linear combination of selected basis functions, and learn this continuous function by exploiting a deep network to predict the linear combination coefficients. Then, the learned latent continuous function is exploited for interpolation in backprojection step, which first time takes the advantage of deep learning for the interpolation in FBP. Extensive experiments, which encompass diverse CT scenarios, demonstrate the effectiveness of the proposed LInFBP in terms of enhanced reconstructed image quality, plug-and-play ability and generalization capability.
SS-CTML: Self-Supervised Cross-Task Mutual Learning for CT Image Reconstruction
Dec 31, 2024



Abstract:Supervised deep-learning (SDL) techniques with paired training datasets have been widely studied for X-ray computed tomography (CT) image reconstruction. However, due to the difficulties of obtaining paired training datasets in clinical routine, the SDL methods are still away from common uses in clinical practices. In recent years, self-supervised deep-learning (SSDL) techniques have shown great potential for the studies of CT image reconstruction. In this work, we propose a self-supervised cross-task mutual learning (SS-CTML) framework for CT image reconstruction. Specifically, a sparse-view scanned and a limited-view scanned sinogram data are first extracted from a full-view scanned sinogram data, which results in three individual reconstruction tasks, i.e., the full-view CT (FVCT) reconstruction, the sparse-view CT (SVCT) reconstruction, and limited-view CT (LVCT) reconstruction. Then, three neural networks are constructed for the three reconstruction tasks. Considering that the ultimate goals of the three tasks are all to reconstruct high-quality CT images, we therefore construct a set of cross-task mutual learning objectives for the three tasks, in which way, the three neural networks can be self-supervised optimized by learning from each other. Clinical datasets are adopted to evaluate the effectiveness of the proposed framework. Experimental results demonstrate that the SS-CTML framework can obtain promising CT image reconstruction performance in terms of both quantitative and qualitative measurements.
Large-vocabulary forensic pathological analyses via prototypical cross-modal contrastive learning
Jul 20, 2024Abstract:Forensic pathology is critical in determining the cause and manner of death through post-mortem examinations, both macroscopic and microscopic. The field, however, grapples with issues such as outcome variability, laborious processes, and a scarcity of trained professionals. This paper presents SongCi, an innovative visual-language model (VLM) designed specifically for forensic pathology. SongCi utilizes advanced prototypical cross-modal self-supervised contrastive learning to enhance the accuracy, efficiency, and generalizability of forensic analyses. It was pre-trained and evaluated on a comprehensive multi-center dataset, which includes over 16 million high-resolution image patches, 2,228 vision-language pairs of post-mortem whole slide images (WSIs), and corresponding gross key findings, along with 471 distinct diagnostic outcomes. Our findings indicate that SongCi surpasses existing multi-modal AI models in many forensic pathology tasks, performs comparably to experienced forensic pathologists and significantly better than less experienced ones, and provides detailed multi-modal explainability, offering critical assistance in forensic investigations. To the best of our knowledge, SongCi is the first VLM specifically developed for forensic pathological analysis and the first large-vocabulary computational pathology (CPath) model that directly processes gigapixel WSIs in forensic science.
DA-TransUNet: Integrating Spatial and Channel Dual Attention with Transformer U-Net for Medical Image Segmentation
Oct 19, 2023Abstract:Great progress has been made in automatic medical image segmentation due to powerful deep representation learning. The influence of transformer has led to research into its variants, and large-scale replacement of traditional CNN modules. However, such trend often overlooks the intrinsic feature extraction capabilities of the transformer and potential refinements to both the model and the transformer module through minor adjustments. This study proposes a novel deep medical image segmentation framework, called DA-TransUNet, aiming to introduce the Transformer and dual attention block into the encoder and decoder of the traditional U-shaped architecture. Unlike prior transformer-based solutions, our DA-TransUNet utilizes attention mechanism of transformer and multifaceted feature extraction of DA-Block, which can efficiently combine global, local, and multi-scale features to enhance medical image segmentation. Meanwhile, experimental results show that a dual attention block is added before the Transformer layer to facilitate feature extraction in the U-net structure. Furthermore, incorporating dual attention blocks in skip connections can enhance feature transfer to the decoder, thereby improving image segmentation performance. Experimental results across various benchmark of medical image segmentation reveal that DA-TransUNet significantly outperforms the state-of-the-art methods. The codes and parameters of our model will be publicly available at https://github.com/SUN-1024/DA-TransUnet.
A Peer-to-peer Federated Continual Learning Network for Improving CT Imaging from Multiple Institutions
Jun 03, 2023



Abstract:Deep learning techniques have been widely used in computed tomography (CT) but require large data sets to train networks. Moreover, data sharing among multiple institutions is limited due to data privacy constraints, which hinders the development of high-performance DL-based CT imaging models from multi-institutional collaborations. Federated learning (FL) strategy is an alternative way to train the models without centralizing data from multi-institutions. In this work, we propose a novel peer-to-peer federated continual learning strategy to improve low-dose CT imaging performance from multiple institutions. The newly proposed method is called peer-to-peer continual FL with intermediate controllers, i.e., icP2P-FL. Specifically, different from the conventional FL model, the proposed icP2P-FL does not require a central server that coordinates training information for a global model. In the proposed icP2P-FL method, the peer-to-peer federated continual learning is introduced wherein the DL-based model is continually trained one client after another via model transferring and inter institutional parameter sharing due to the common characteristics of CT data among the clients. Furthermore, an intermediate controller is developed to make the overall training more flexible. Numerous experiments were conducted on the AAPM low-dose CT Grand Challenge dataset and local datasets, and the experimental results showed that the proposed icP2P-FL method outperforms the other comparative methods both qualitatively and quantitatively, and reaches an accuracy similar to a model trained with pooling data from all the institutions.
Lesion classification by model-based feature extraction: A differential affine invariant model of soft tissue elasticity
May 27, 2022
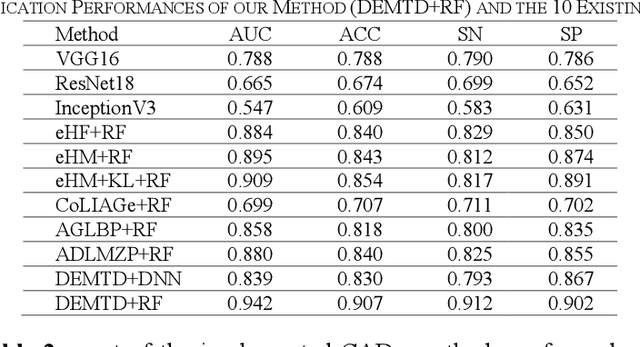
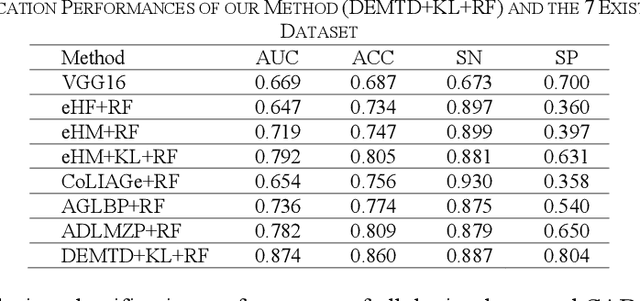
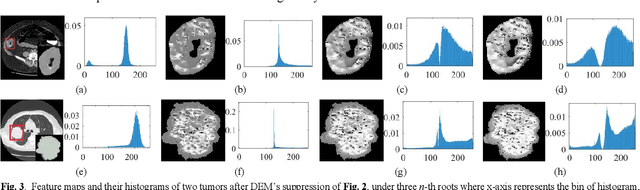
Abstract:The elasticity of soft tissues has been widely considered as a characteristic property to differentiate between healthy and vicious tissues and, therefore, motivated several elasticity imaging modalities, such as Ultrasound Elastography, Magnetic Resonance Elastography, and Optical Coherence Elastography. This paper proposes an alternative approach of modeling the elasticity using Computed Tomography (CT) imaging modality for model-based feature extraction machine learning (ML) differentiation of lesions. The model describes a dynamic non-rigid (or elastic) deformation in differential manifold to mimic the soft tissues elasticity under wave fluctuation in vivo. Based on the model, three local deformation invariants are constructed by two tensors defined by the first and second order derivatives from the CT images and used to generate elastic feature maps after normalization via a novel signal suppression method. The model-based elastic image features are extracted from the feature maps and fed to machine learning to perform lesion classifications. Two pathologically proven image datasets of colon polyps (44 malignant and 43 benign) and lung nodules (46 malignant and 20 benign) were used to evaluate the proposed model-based lesion classification. The outcomes of this modeling approach reached the score of area under the curve of the receiver operating characteristics of 94.2 % for the polyps and 87.4 % for the nodules, resulting in an average gain of 5 % to 30 % over ten existing state-of-the-art lesion classification methods. The gains by modeling tissue elasticity for ML differentiation of lesions are striking, indicating the great potential of exploring the modeling strategy to other tissue properties for ML differentiation of lesions.
Direct Energy-resolving CT Imaging via Energy-integrating CT images using a Unified Generative Adversarial Network
Oct 14, 2019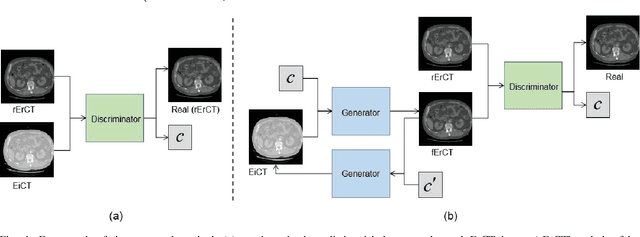
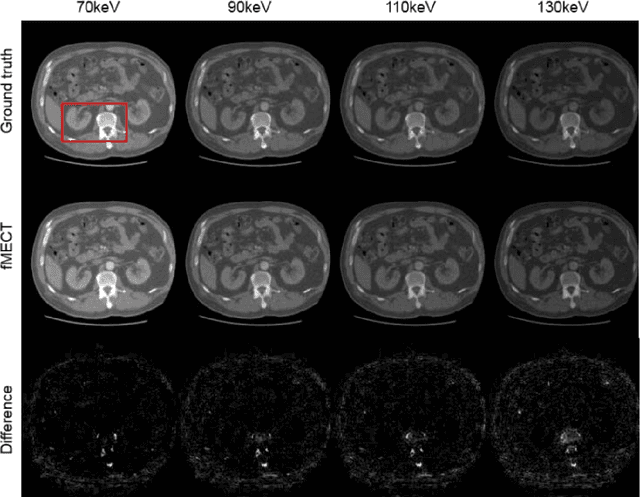
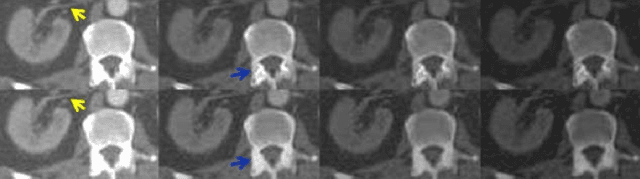
Abstract:Energy-resolving computed tomography (ErCT) has the ability to acquire energy-dependent measurements simultaneously and quantitative material information with improved contrast-to-noise ratio. Meanwhile, ErCT imaging system is usually equipped with an advanced photon counting detector, which is expensive and technically complex. Therefore, clinical ErCT scanners are not yet commercially available, and they are in various stage of completion. This makes the researchers less accessible to the ErCT images. In this work, we investigate to produce ErCT images directly from existing energy-integrating CT (EiCT) images via deep neural network. Specifically, different from other networks that produce ErCT images at one specific energy, this model employs a unified generative adversarial network (uGAN) to concurrently train EiCT datasets and ErCT datasets with different energies and then performs image-to-image translation from existing EiCT images to multiple ErCT image outputs at various energy bins. In this study, the present uGAN generates ErCT images at 70keV, 90keV, 110keV, and 130keV simultaneously from EiCT images at140kVp. We evaluate the present uGAN model on a set of over 1380 CT image slices and show that the present uGAN model can produce promising ErCT estimation results compared with the ground truth qualitatively and quantitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge