Jan Kirschke
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025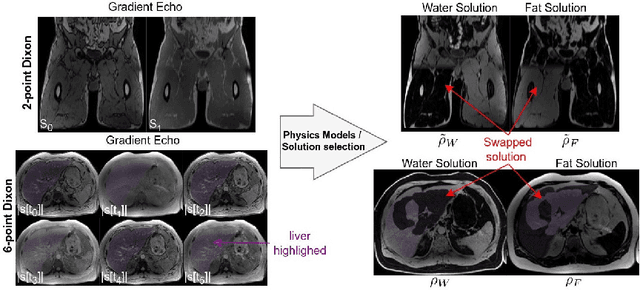
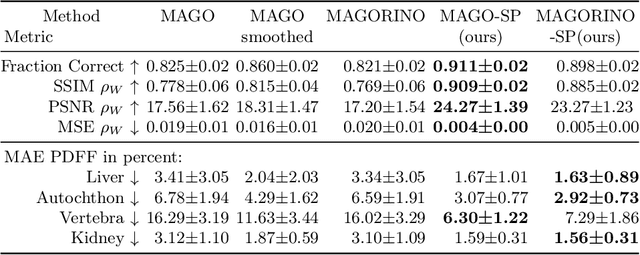
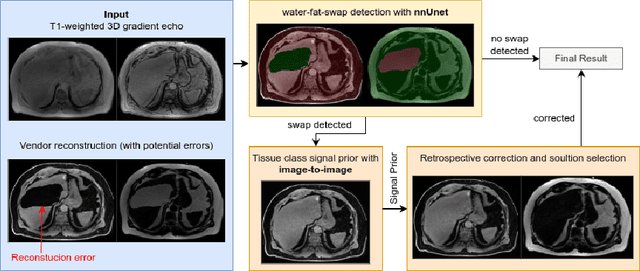
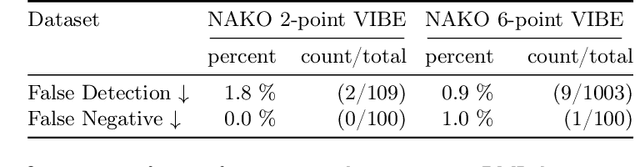
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
PARASIDE: An Automatic Paranasal Sinus Segmentation and Structure Analysis Tool for MRI
Jan 24, 2025



Abstract:Chronic rhinosinusitis (CRS) is a common and persistent sinus imflammation that affects 5 - 12\% of the general population. It significantly impacts quality of life and is often difficult to assess due to its subjective nature in clinical evaluation. We introduce PARASIDE, an automatic tool for segmenting air and soft tissue volumes of the structures of the sinus maxillaris, frontalis, sphenodalis and ethmoidalis in T1 MRI. By utilizing that segmentation, we can quantify feature relations that have been observed only manually and subjectively before. We performed an exemplary study and showed both volume and intensity relations between structures and radiology reports. While the soft tissue segmentation is good, the automated annotations of the air volumes are excellent. The average intensity over air structures are consistently below those of the soft tissues, close to perfect separability. Healthy subjects exhibit lower soft tissue volumes and lower intensities. Our developed system is the first automated whole nasal segmentation of 16 structures, and capable of calculating medical relevant features such as the Lund-Mackay score.
Enhancing Interpretability of Vertebrae Fracture Grading using Human-interpretable Prototypes
Apr 03, 2024



Abstract:Vertebral fracture grading classifies the severity of vertebral fractures, which is a challenging task in medical imaging and has recently attracted Deep Learning (DL) models. Only a few works attempted to make such models human-interpretable despite the need for transparency and trustworthiness in critical use cases like DL-assisted medical diagnosis. Moreover, such models either rely on post-hoc methods or additional annotations. In this work, we propose a novel interpretable-by-design method, ProtoVerse, to find relevant sub-parts of vertebral fractures (prototypes) that reliably explain the model's decision in a human-understandable way. Specifically, we introduce a novel diversity-promoting loss to mitigate prototype repetitions in small datasets with intricate semantics. We have experimented with the VerSe'19 dataset and outperformed the existing prototype-based method. Further, our model provides superior interpretability against the post-hoc method. Importantly, expert radiologists validated the visual interpretability of our results, showing clinical applicability.
The Brain Tumor Segmentation Challenge 2023: Brain MR Image Synthesis for Tumor Segmentation
May 20, 2023
Abstract:Automated brain tumor segmentation methods are well established, reaching performance levels with clear clinical utility. Most algorithms require four input magnetic resonance imaging (MRI) modalities, typically T1-weighted images with and without contrast enhancement, T2-weighted images, and FLAIR images. However, some of these sequences are often missing in clinical practice, e.g., because of time constraints and/or image artifacts (such as patient motion). Therefore, substituting missing modalities to recover segmentation performance in these scenarios is highly desirable and necessary for the more widespread adoption of such algorithms in clinical routine. In this work, we report the set-up of the Brain MR Image Synthesis Benchmark (BraSyn), organized in conjunction with the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2023. The objective of the challenge is to benchmark image synthesis methods that realistically synthesize missing MRI modalities given multiple available images to facilitate automated brain tumor segmentation pipelines. The image dataset is multi-modal and diverse, created in collaboration with various hospitals and research institutions.
The Brain Tumor Segmentation Challenge 2023: Local Synthesis of Healthy Brain Tissue via Inpainting
May 15, 2023



Abstract:A myriad of algorithms for the automatic analysis of brain MR images is available to support clinicians in their decision-making. For brain tumor patients, the image acquisition time series typically starts with a scan that is already pathological. This poses problems, as many algorithms are designed to analyze healthy brains and provide no guarantees for images featuring lesions. Examples include but are not limited to algorithms for brain anatomy parcellation, tissue segmentation, and brain extraction. To solve this dilemma, we introduce the BraTS 2023 inpainting challenge. Here, the participants' task is to explore inpainting techniques to synthesize healthy brain scans from lesioned ones. The following manuscript contains the task formulation, dataset, and submission procedure. Later it will be updated to summarize the findings of the challenge. The challenge is organized as part of the BraTS 2023 challenge hosted at the MICCAI 2023 conference in Vancouver, Canada.
Approaching Peak Ground Truth
Dec 31, 2022

Abstract:Machine learning models are typically evaluated by computing similarity with reference annotations and trained by maximizing similarity with such. Especially in the bio-medical domain, annotations are subjective and suffer from low inter- and intra-rater reliability. Since annotations only reflect the annotation entity's interpretation of the real world, this can lead to sub-optimal predictions even though the model achieves high similarity scores. Here, the theoretical concept of Peak Ground Truth (PGT) is introduced. PGT marks the point beyond which an increase in similarity with the reference annotation stops translating to better Real World Model Performance (RWMP). Additionally, a quantitative technique to approximate PGT by computing inter- and intra-rater reliability is proposed. Finally, three categories of PGT-aware strategies to evaluate and improve model performance are reviewed.
CheXplaining in Style: Counterfactual Explanations for Chest X-rays using StyleGAN
Jul 15, 2022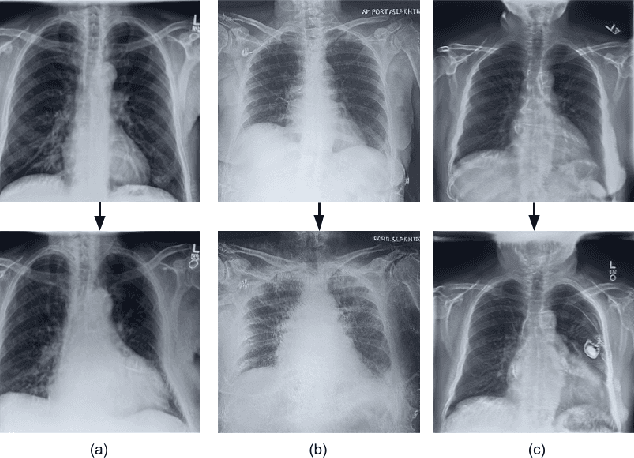
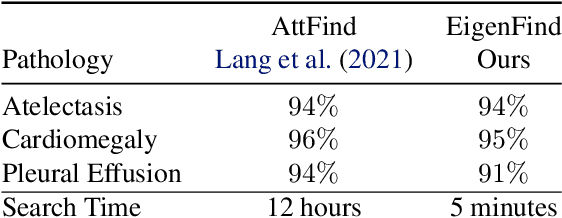
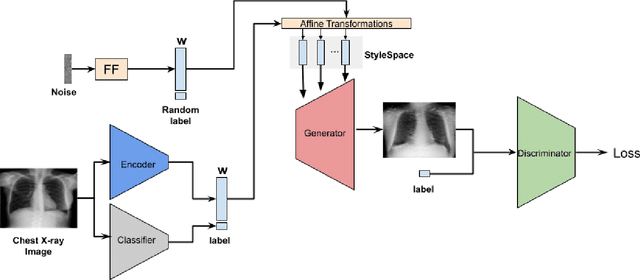
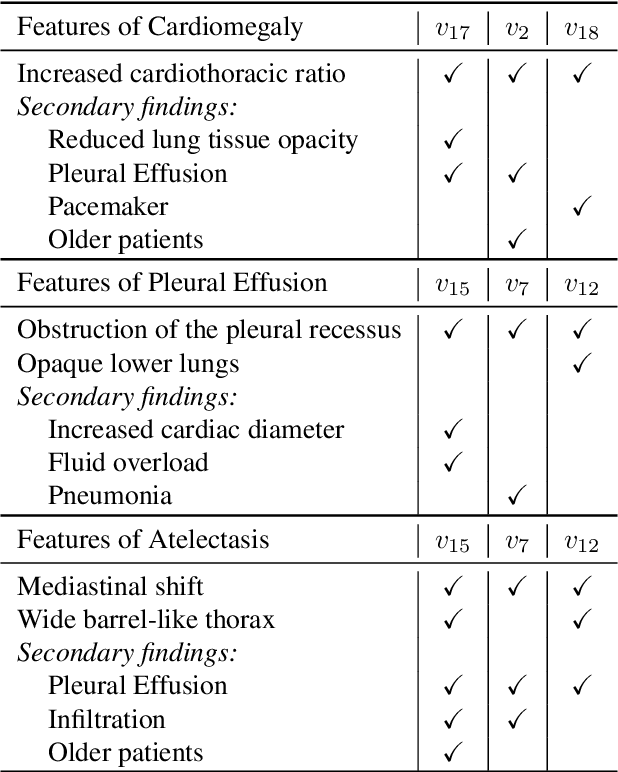
Abstract:Deep learning models used in medical image analysis are prone to raising reliability concerns due to their black-box nature. To shed light on these black-box models, previous works predominantly focus on identifying the contribution of input features to the diagnosis, i.e., feature attribution. In this work, we explore counterfactual explanations to identify what patterns the models rely on for diagnosis. Specifically, we investigate the effect of changing features within chest X-rays on the classifier's output to understand its decision mechanism. We leverage a StyleGAN-based approach (StyleEx) to create counterfactual explanations for chest X-rays by manipulating specific latent directions in their latent space. In addition, we propose EigenFind to significantly reduce the computation time of generated explanations. We clinically evaluate the relevancy of our counterfactual explanations with the help of radiologists. Our code is publicly available.
Deep Quality Estimation: Creating Surrogate Models for Human Quality Ratings
May 17, 2022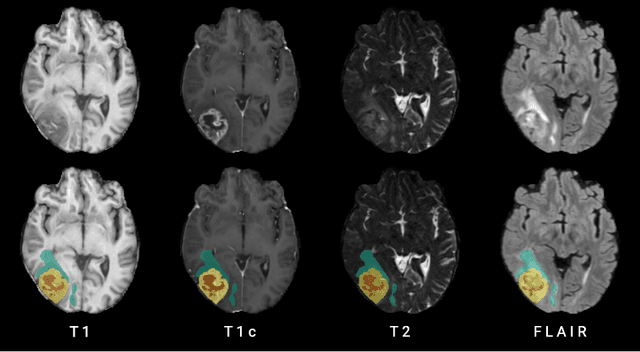
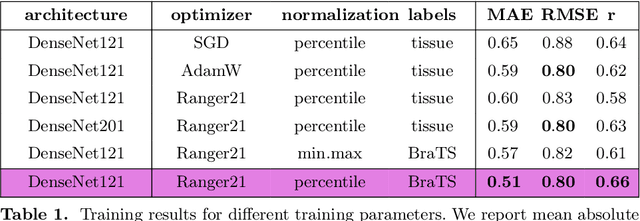
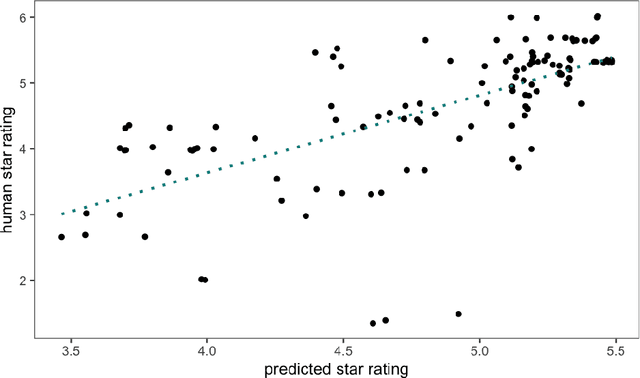
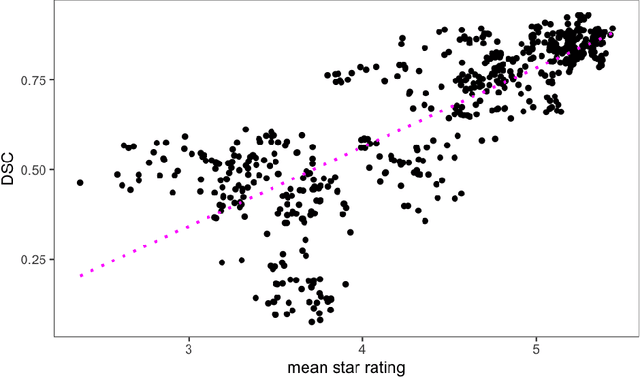
Abstract:Human ratings are abstract representations of segmentation quality. To approximate human quality ratings on scarce expert data, we train surrogate quality estimation models. We evaluate on a complex multi-class segmentation problem, specifically glioma segmentation following the BraTS annotation protocol. The training data features quality ratings from 15 expert neuroradiologists on a scale ranging from 1 to 6 stars for various computer-generated and manual 3D annotations. Even though the networks operate on 2D images and with scarce training data, we can approximate segmentation quality within a margin of error comparable to human intra-rater reliability. Segmentation quality prediction has broad applications. While an understanding of segmentation quality is imperative for successful clinical translation of automatic segmentation quality algorithms, it can play an essential role in training new segmentation models. Due to the split-second inference times, it can be directly applied within a loss function or as a fully-automatic dataset curation mechanism in a federated learning setting.
blob loss: instance imbalance aware loss functions for semantic segmentation
May 17, 2022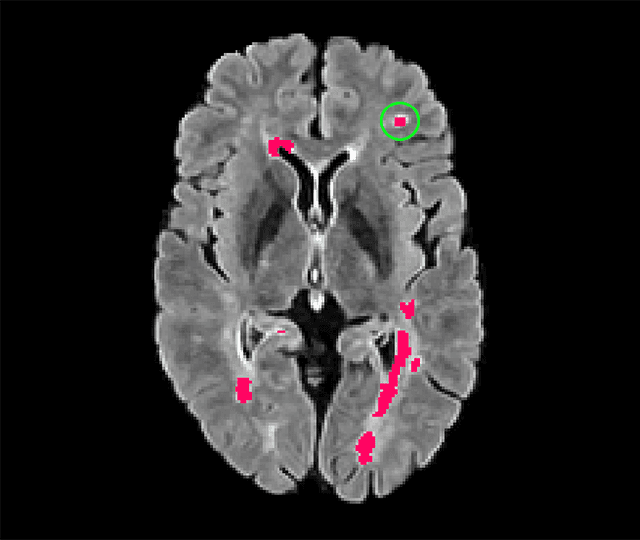
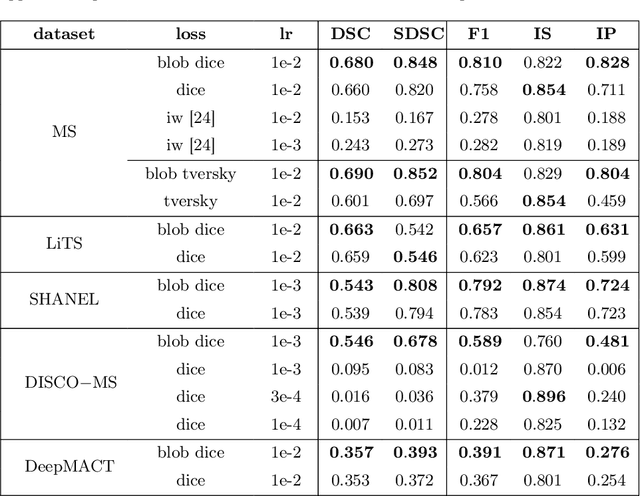

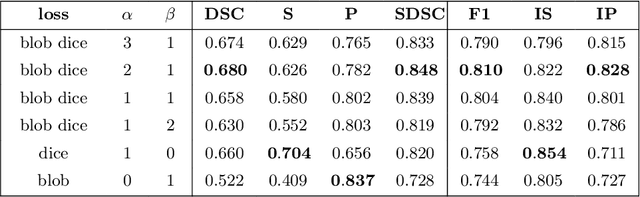
Abstract:Deep convolutional neural networks have proven to be remarkably effective in semantic segmentation tasks. Most popular loss functions were introduced targeting improved volumetric scores, such as the Sorensen Dice coefficient. By design, DSC can tackle class imbalance; however, it does not recognize instance imbalance within a class. As a result, a large foreground instance can dominate minor instances and still produce a satisfactory Sorensen Dice coefficient. Nevertheless, missing out on instances will lead to poor detection performance. This represents a critical issue in applications such as disease progression monitoring. For example, it is imperative to locate and surveil small-scale lesions in the follow-up of multiple sclerosis patients. We propose a novel family of loss functions, nicknamed blob loss, primarily aimed at maximizing instance-level detection metrics, such as F1 score and sensitivity. Blob loss is designed for semantic segmentation problems in which the instances are the connected components within a class. We extensively evaluate a DSC-based blob loss in five complex 3D semantic segmentation tasks featuring pronounced instance heterogeneity in terms of texture and morphology. Compared to soft Dice loss, we achieve 5 percent improvement for MS lesions, 3 percent improvement for liver tumor, and an average 2 percent improvement for Microscopy segmentation tasks considering F1 score.
A Deep Learning Approach to Predicting Collateral Flow in Stroke Patients Using Radiomic Features from Perfusion Images
Oct 24, 2021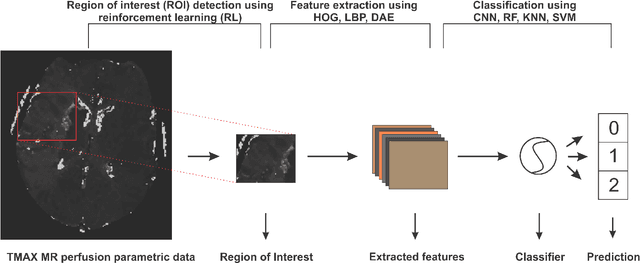
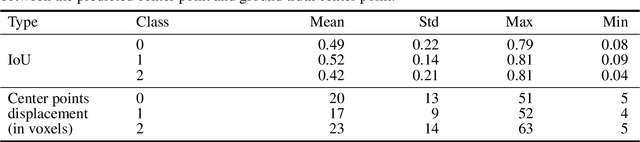
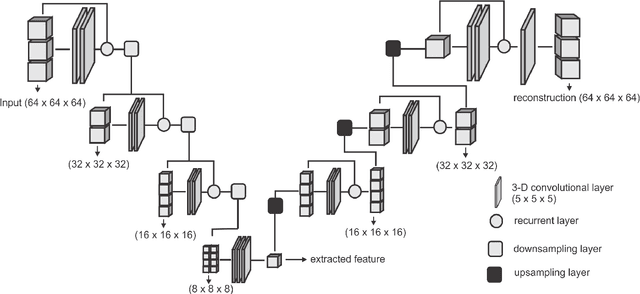
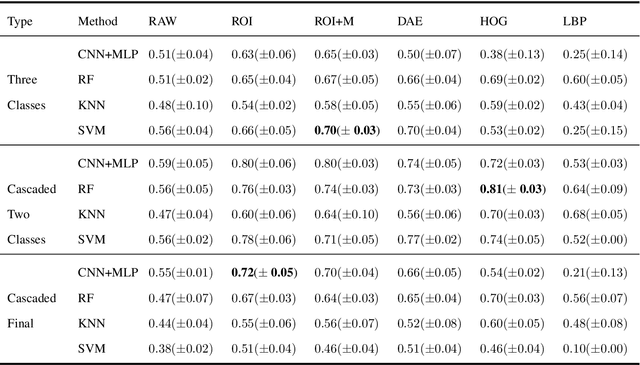
Abstract:Collateral circulation results from specialized anastomotic channels which are capable of providing oxygenated blood to regions with compromised blood flow caused by ischemic injuries. The quality of collateral circulation has been established as a key factor in determining the likelihood of a favorable clinical outcome and goes a long way to determine the choice of stroke care model - that is the decision to transport or treat eligible patients immediately. Though there exist several imaging methods and grading criteria for quantifying collateral blood flow, the actual grading is mostly done through manual inspection of the acquired images. This approach is associated with a number of challenges. First, it is time-consuming - the clinician needs to scan through several slices of images to ascertain the region of interest before deciding on what severity grade to assign to a patient. Second, there is a high tendency for bias and inconsistency in the final grade assigned to a patient depending on the experience level of the clinician. We present a deep learning approach to predicting collateral flow grading in stroke patients based on radiomic features extracted from MR perfusion data. First, we formulate a region of interest detection task as a reinforcement learning problem and train a deep learning network to automatically detect the occluded region within the 3D MR perfusion volumes. Second, we extract radiomic features from the obtained region of interest through local image descriptors and denoising auto-encoders. Finally, we apply a convolutional neural network and other machine learning classifiers to the extracted radiomic features to automatically predict the collateral flow grading of the given patient volume as one of three severity classes - no flow (0), moderate flow (1), and good flow (2)...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge